3ZDT
 
 | | Crystal structure of basic patch mutant FAK FERM domain FAK31- 405 K216A, K218A, R221A, K222A | | Descriptor: | FOCAL ADHESION KINASE 1 | | Authors: | Goni, G.M, Epifano, C, Boskovic, J, Camacho-Artacho, M, Zhou, J, Martin, M.T, Eck, M.J, Kremer, L, Graeter, F, Gervasio, F.L, Perez-Moreno, M, Lietha, D. | | Deposit date: | 2012-11-30 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Phosphatidylinositol 4,5-Bisphosphate Triggers Activation of Focal Adhesion Kinase by Inducing Clustering and Conformational Changes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4CYE
 
 | | Crystal structure of avian FAK FERM domain FAK31-405 at 3.2A | | Descriptor: | FOCAL ADHESION KINASE 1 | | Authors: | Goni, G.M, Epifano, C, Boskovic, J, Camacho-Artacho, M, Zhou, J, Martin, M.T, Eck, M.J, Kremer, L, Graeter, F, Gervasio, F.L, Perez-Moreno, M, Lietha, D. | | Deposit date: | 2014-04-10 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Phosphatidylinositol 4,5-Bisphosphate Triggers Activation of Focal Adhesion Kinase by Inducing Clustering and Conformational Changes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6TY3
 
 | | FAK structure from single particle analysis of 2D crystals | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Acebron, I, Righetto, R, Biyani, N, Chami, M, Boskovic, J, Stahlberg, H, Lietha, D. | | Deposit date: | 2020-01-15 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.32 Å) | | Cite: | Structural basis of Focal Adhesion Kinase activation on lipid membranes.
Embo J., 39, 2020
|
|
6TY4
 
 | | FAK structure with AMP-PNP from single particle analysis of 2D crystals | | Descriptor: | Focal adhesion kinase 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Acebron, I, Righetto, R, Biyani, N, Chami, M, Boskovic, J, Stahlberg, H, Lietha, D. | | Deposit date: | 2020-01-15 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.96 Å) | | Cite: | Structural basis of Focal Adhesion Kinase activation on lipid membranes.
Embo J., 39, 2020
|
|
7B00
 
 | | Human LAT2-4F2hc complex in the apo-state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Digitonin, Isoform 2 of 4F2 cell-surface antigen heavy chain, ... | | Authors: | Rodriguez, C.F, Escudero-Bravo, P, Garcia-Martin, C, Boskovic, J, Errasti-Murugarren, E, Palacin, M, Llorca, O. | | Deposit date: | 2020-11-17 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structural basis for substrate specificity of heteromeric transporters of neutral amino acids.
Proc Natl Acad Sci U S A, 118, 2021
|
|
5NNQ
 
 | |
5NNN
 
 | |
9EQ2
 
 | | Arabidopsis thaliana R2T complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, At1g56440, RuvB-like helicase, ... | | Authors: | Lopez-Perrote, A, Llorca, O. | | Deposit date: | 2024-03-20 | | Release date: | 2025-01-29 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | The structure of the R2T complex reveals a different architecture from the related HSP90 cochaperone R2TP.
Structure, 33, 2025
|
|
7AHO
 
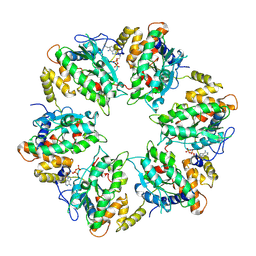 | | RUVBL1-RUVBL2 heterohexameric ring after binding of RNA helicase DHX34 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2 | | Authors: | Lopez-Perrote, A, Rodriguez, C.F, Llorca, O. | | Deposit date: | 2020-09-25 | | Release date: | 2020-11-25 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Regulation of RUVBL1-RUVBL2 AAA-ATPases by the nonsense-mediated mRNA decay factor DHX34, as evidenced by Cryo-EM.
Elife, 9, 2020
|
|
9EMA
 
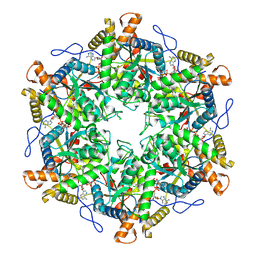 | | RUVBL1/2 in complex with ATP and CB-6644 inhibitor | | Descriptor: | 5-chloranyl-2-ethoxy-4-fluoranyl-~{N}-[4-[[3-(methoxymethyl)-1-oxidanylidene-6,7-dihydro-5~{H}-pyrazolo[1,2-a][1,2]benzodiazepin-2-yl]amino]-2,2-dimethyl-4-oxidanylidene-butyl]benzamide, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lopez-Perrote, A, Llorca, O, Garcia-Martin, C. | | Deposit date: | 2024-03-07 | | Release date: | 2024-05-15 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Mechanism of allosteric inhibition of RUVBL1-RUVBL2 by the small-molecule CB-6644
Cell Rep Phys Sci, 2024
|
|
9EMC
 
 | |
8AG3
 
 | |
8AG5
 
 | | Vaccinia C16 protein bound to Ku70/Ku80 | | Descriptor: | Ku70-Xrcc6, Protein C10, X-ray repair cross-complementing protein 5 | | Authors: | Rivera-Calzada, A, Arribas-Bosacoma, R, Pearl, L.H, Llorca, O. | | Deposit date: | 2022-07-19 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural basis for the inactivation of cytosolic DNA sensing by the vaccinia virus.
Nat Commun, 13, 2022
|
|
8AG4
 
 | | Vaccinia C16 protein bound to Ku70/Ku80 | | Descriptor: | Protein C10, X-ray repair cross-complementing protein 5, X-ray repair cross-complementing protein 6 | | Authors: | Rivera-Calzada, A, Arribas-Bosacoma, R, Pearl, L.H, Llorca, O. | | Deposit date: | 2022-07-19 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Structural basis for the inactivation of cytosolic DNA sensing by the vaccinia virus.
Nat Commun, 13, 2022
|
|
5NNL
 
 | |
5A52
 
 | |
5A51
 
 | |
5A4X
 
 | |
5A50
 
 | |
