3CYJ
 
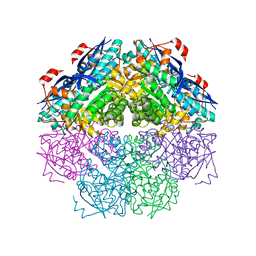 | | Crystal structure of a mandelate racemase/muconate lactonizing enzyme-like protein from Rubrobacter xylanophilus | | Descriptor: | GLYCEROL, Mandelate racemase/muconate lactonizing enzyme-like protein, SODIUM ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Zhang, F, Bravo, J, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a mandelate racemase/muconate lactonizing enzyme-like protein from Rubrobacter xylanophilus.
To be Published
|
|
3CTA
 
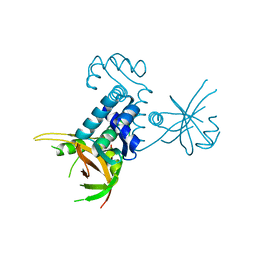 | | Crystal structure of riboflavin kinase from Thermoplasma acidophilum | | Descriptor: | Riboflavin kinase | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Mendoza, M, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-11 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of riboflavin kinase from Thermoplasma acidophilum.
To be Published
|
|
3SN1
 
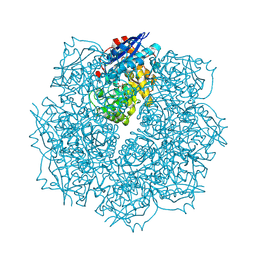 | | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium and tartrate | | Descriptor: | CHLORIDE ION, D(-)-TARTARIC ACID, MAGNESIUM ION, ... | | Authors: | Bonanno, J.B, Patskovsky, Y, Toro, R, Dickey, M, Bain, K.T, Wu, B, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium and tartrate
To be Published
|
|
3SN4
 
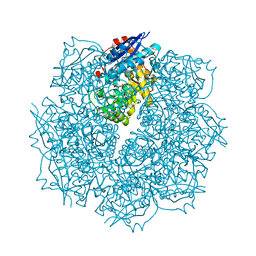 | | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium and alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bonanno, J.B, Patskovsky, Y, Toro, R, Dickey, M, Bain, K.T, Wu, B, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium and alpha-ketoglutarate
To be Published
|
|
3NIW
 
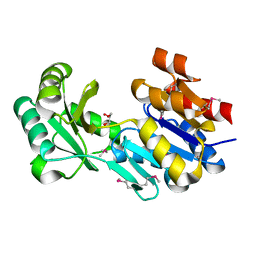 | | Crystal structure of a haloacid dehalogenase-like hydrolase from Bacteroides thetaiotaomicron | | Descriptor: | GLYCEROL, Haloacid dehalogenase-like hydrolase, MAGNESIUM ION | | Authors: | Bonanno, J.B, Ramagopal, U, Toro, R, Rutter, M, Bain, K.T, Wu, B, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-16 | | Release date: | 2010-06-30 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a haloacid dehalogenase-like hydrolase from Bacteroides thetaiotaomicron
To be Published
|
|
3SN0
 
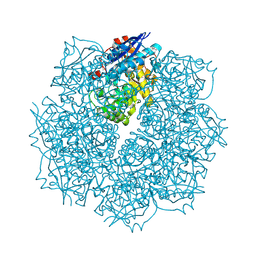 | | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium and fumarate | | Descriptor: | CHLORIDE ION, FUMARIC ACID, MAGNESIUM ION, ... | | Authors: | Bonanno, J.B, Patskovsky, Y, Toro, R, Dickey, M, Bain, K.T, Wu, B, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium and fumarate
To be Published
|
|
1FI4
 
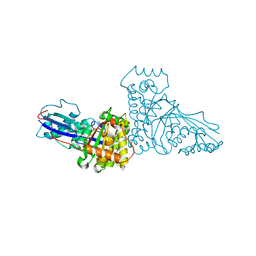 | | THE X-RAY CRYSTAL STRUCTURE OF MEVALONATE 5-DIPHOSPHATE DECARBOXYLASE AT 2.3 ANGSTROM RESOLUTION. | | Descriptor: | MEVALONATE 5-DIPHOSPHATE DECARBOXYLASE | | Authors: | Bonanno, J.B, Edo, C, Eswar, N, Pieper, U, Romanowski, M.J, Ilyin, V, Gerchman, S.E, Kycia, H, Studier, F.W, Sali, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2000-08-03 | | Release date: | 2001-03-21 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural genomics of enzymes involved in sterol/isoprenoid biosynthesis.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1Q2W
 
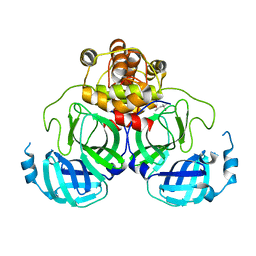 | | X-Ray Crystal Structure of the SARS Coronavirus Main Protease | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3C-like protease | | Authors: | Bonanno, J.B, Fowler, R, Gupta, S, Hendle, J, Lorimer, D, Romero, R, Sauder, J.M, Wei, C.L, Liu, E.T, Burley, S.K, Harris, T. | | Deposit date: | 2003-07-26 | | Release date: | 2003-07-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Company Says It Mapped Part of SARS Virus
New York Times, 30 July, 2003
|
|
3B40
 
 | | Crystal structure of the probable dipeptidase PvdM from Pseudomonas aeruginosa | | Descriptor: | CADMIUM ION, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Bonanno, J.B, Patskovsky, Y, Dickey, M, Bain, K.T, Mendoza, M, Fong, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-23 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the probable dipeptidase PvdM from Pseudomonas aeruginosa.
To be Published
|
|
3BGH
 
 | | Crystal structure of putative neuraminyllactose-binding hemagglutinin homolog from Helicobacter pylori | | Descriptor: | Putative neuraminyllactose-binding hemagglutinin homolog, SULFATE ION | | Authors: | Bonanno, J.B, Dickey, J, Bain, K.T, McKenzie, C, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-11-26 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of putative neuraminyllactose-binding hemagglutinin homolog from Helicobacter pylori.
To be Published
|
|
3BHW
 
 | | Crystal structure of an uncharacterized protein from Magnetospirillum magneticum | | Descriptor: | Uncharacterized protein | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Lau, C, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-11-29 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of an uncharacterized protein from Magnetospirillum magneticum.
To be Published
|
|
3BIL
 
 | | Crystal structure of a probable LacI family transcriptional regulator from Corynebacterium glutamicum | | Descriptor: | Probable LacI-family transcriptional regulator | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Mendoza, M, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a probable LacI family transcriptional regulator from Corynebacterium glutamicum.
To be Published
|
|
3BUT
 
 | | Crystal structure of protein Af_0446 from Archaeoglobus fulgidus | | Descriptor: | Uncharacterized protein Af_0446 | | Authors: | Bonanno, J.B, Patskovsky, Y, Ozyurt, S, Ashok, S, Zhang, F, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-03 | | Release date: | 2008-01-15 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of protein Af_0446 from Archaeoglobus fulgidus.
To be Published
|
|
3C8T
 
 | | Crystal structure of fumarate lyase from Mesorhizobium sp. BNC1 | | Descriptor: | Fumarate lyase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-13 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of fumarate lyase from Mesorhizobium sp. BNC1.
To be Published
|
|
3BS4
 
 | | Crystal structure of uncharacterized protein PH0321 from Pyrococcus horikoshii in complex with an unknown peptide | | Descriptor: | Uncharacterized protein PH0321, Unknown peptide | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Hu, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-21 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of uncharacterized protein PH0321 from Pyrococcus horikoshii in complex with an unknown peptide.
To be Published
|
|
3C97
 
 | | Crystal structure of the response regulator receiver domain of a signal transduction histidine kinase from Aspergillus oryzae | | Descriptor: | Signal transduction histidine kinase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-15 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the response regulator receiver domain of a signal transduction histidine kinase from Aspergillus oryzae.
To be Published
|
|
3CBW
 
 | | Crystal structure of the YdhT protein from Bacillus subtilis | | Descriptor: | CITRIC ACID, YdhT protein | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Iizuka, M, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-23 | | Release date: | 2008-03-11 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.269 Å) | | Cite: | Crystal structure of the YdhT protein from Bacillus subtilis.
To be Published
|
|
3CDX
 
 | | Crystal structure of succinylglutamatedesuccinylase/aspartoacylase from Rhodobacter sphaeroides | | Descriptor: | CALCIUM ION, Succinylglutamatedesuccinylase/aspartoacylase | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Iizuka, M, Patterson, K, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-27 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of succinylglutamatedesuccinylase/aspartoacylase from Rhodobacter sphaeroides.
To be Published
|
|
3CTD
 
 | | Crystal structure of a putative AAA family ATPase from Prochlorococcus marinus subsp. pastoris | | Descriptor: | Putative ATPase, AAA family | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Lau, C, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-11 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative AAA family ATPase from Prochlorococcus marinus subsp. pastoris.
To be Published
|
|
3CLW
 
 | | Crystal structure of conserved exported protein from Bacteroides fragilis | | Descriptor: | Conserved exported protein | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Chang, S, Ozyurt, K, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-20 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Crystal structure of conserved exported protein from Bacteroides fragilis.
To be Published
|
|
3D19
 
 | | Crystal structure of a conserved metalloprotein from Bacillus cereus | | Descriptor: | Conserved metalloprotein, FE (III) ION, MAGNESIUM ION | | Authors: | Bonanno, J.B, Patskovsky, Y, Freeman, J, Bain, K.T, Chang, S, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-05-05 | | Release date: | 2008-07-08 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a conserved metalloprotein from Bacillus cereus.
To be Published
|
|
1I9A
 
 | | STRUCTURAL STUDIES OF CHOLESTEROL BIOSYNTHESIS: MEVALONATE 5-DIPHOSPHATE DECARBOXYLASE AND ISOPENTENYL DIPHOSPHATE ISOMERASE | | Descriptor: | ISOPENTENYL-DIPHOSPHATE DELTA-ISOMERASE, MANGANESE (II) ION | | Authors: | Bonanno, J.B, Edo, C, Eswar, N, Pieper, U, Romanowski, M.J, Ilyin, V, Gerchman, S.E, Kycia, H, Studier, F.W, Sali, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-03-18 | | Release date: | 2001-03-28 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural genomics of enzymes involved in sterol/isoprenoid biosynthesis.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3I6V
 
 | | Crystal structure of a periplasmic His/Glu/Gln/Arg/opine family-binding protein from Silicibacter pomeroyi in complex with lysine | | Descriptor: | GLYCEROL, LYSINE, SODIUM ION, ... | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Miller, S, Romero, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-07 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a periplasmic His/Glu/Gln/Arg/opine family-binding protein from Silicibacter pomeroyi in complex with lysine
To be Published
|
|
3IB6
 
 | | Crystal structure of an uncharacterized protein from Listeria monocytogenes serotype 4b | | Descriptor: | Uncharacterized protein | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Do, J, Ozyurt, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-15 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an uncharacterized protein from Listeria monocytogenes serotype 4b
To be Published
|
|
3IJD
 
 | | Uncharacterized protein Cthe_2304 from Clostridium thermocellum binds two copies of 5-methyl-5,6,7,8-tetrahydrofolic acid | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, uncharacterized protein | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Miller, S, Ozyurt, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-04 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Uncharacterized protein Cthe_2304 from Clostridium thermocellum binds two copies of 5-methyl-5,6,7,8-tetrahydrofolic acid
To be Published
|
|
