1UR6
 
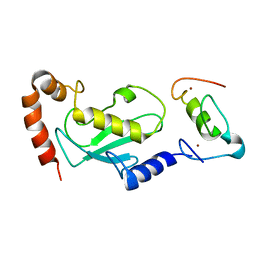 | | NMR based structural model of the UbcH5B-CNOT4 complex | | Descriptor: | POTENTIAL TRANSCRIPTIONAL REPRESSOR NOT4HP, UBIQUITIN-CONJUGATING ENZYME E2-17 KDA 2, ZINC ION | | Authors: | Dominguez, C, Bonvin, A.M.J.J, Winkler, G.S, Van Schaik, F.M.A, Timmers, H.Th.M, Boelens, R. | | Deposit date: | 2003-10-27 | | Release date: | 2004-05-07 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR, THEORETICAL MODEL | | Cite: | Structural Model of the Ubch5B/Cnot4 Complex Revealed by Combining NMR, Mutagenesis, and Docking Approaches.
Structure, 12, 2004
|
|
1W4U
 
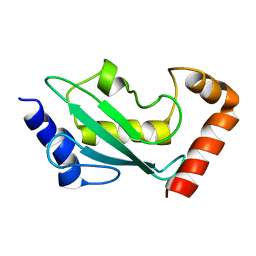 | | NMR solution structure of the ubiquitin conjugating enzyme UbcH5B | | Descriptor: | UBIQUITIN-CONJUGATING ENZYME E2-17 KDA 2 | | Authors: | Houben, K, Dominguez, C, Van Schaik, F.M.A, Timmers, H.T.M, Bonvin, A.M.J.J, Boelens, R. | | Deposit date: | 2004-07-29 | | Release date: | 2004-11-10 | | Last modified: | 2017-04-19 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Ubiquitin-Conjugating Enzyme Ubch5B
J.Mol.Biol., 344, 2004
|
|
2PHE
 
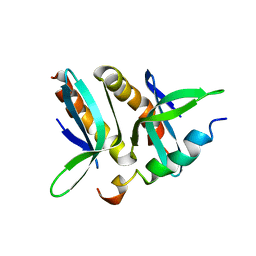 | | Model for VP16 binding to PC4 | | Descriptor: | Alpha trans-inducing protein, TRANSCRIPTIONAL COACTIVATOR PC4 | | Authors: | Jonker, H.R.A, Wechselberger, R.W, Boelens, R, Folkers, G.E, Kaptein, R. | | Deposit date: | 2007-04-11 | | Release date: | 2007-04-24 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structural Properties of the Promiscuous VP16 Activation Domain
Biochemistry, 44, 2005
|
|
2PHG
 
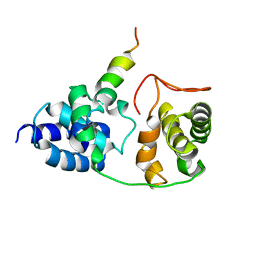 | | Model for VP16 binding to TFIIB | | Descriptor: | Alpha trans-inducing protein, Transcription initiation factor IIB | | Authors: | Jonker, H.R.A, Wechselberger, R.W, Boelens, R, Folkers, G.E, Kaptein, R. | | Deposit date: | 2007-04-11 | | Release date: | 2007-04-24 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structural Properties of the Promiscuous VP16 Activation Domain
Biochemistry, 44, 2005
|
|
1DZ7
 
 | | Solution structure of the a-subunit of human chorionic gonadotropin [modeled without carbohydrate residues] | | Descriptor: | CHORIONIC GONADOTROPIN | | Authors: | Erbel, P.J.A, Karimi-Nejad, Y, De Beer, T, Boelens, R, Kamerling, J.P, Vliegenthart, J.F.G. | | Deposit date: | 2000-02-18 | | Release date: | 2000-02-29 | | Last modified: | 2019-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the alpha-subunit of human chorionic gonadotropin.
Eur.J.Biochem., 260, 1999
|
|
1E9J
 
 | | SOLUTION STRUCTURE OF THE A-SUBUNIT OF HUMAN CHORIONIC GONADOTROPIN [INCLUDING A SINGLE GLCNAC RESIDUE AT ASN52 AND ASN78] | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHORIONIC GONADOTROPIN | | Authors: | Erbel, P.J.A, Karimi-Nejad, Y, van Kuik, J.A, Boelens, R, Kamerling, J.P, Vliegenthart, J.F.G. | | Deposit date: | 2000-10-18 | | Release date: | 2002-07-25 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Effects of the N-linked glycans on the 3D structure of the free alpha-subunit of human chorionic gonadotropin.
Biochemistry, 39, 2000
|
|
1E4U
 
 | | N-terminal RING finger domain of human NOT-4 | | Descriptor: | TRANSCRIPTIONAL REPRESSOR NOT4, ZINC ION | | Authors: | Hanzawa, H, De Ruwe, M.J, Albert, T.K, Van Der Vliet, P.C, Timmers, H.T, Boelens, R. | | Deposit date: | 2000-07-12 | | Release date: | 2001-03-31 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The Structure of the C4C4 Ring Finger of Human not4 Reveals Features Distinct from Those of C3Hc4 Ring Fingers
J.Biol.Chem., 276, 2001
|
|
1HD4
 
 | | SOLUTION STRUCTURE OF THE A-SUBUNIT OF HUMAN CHORIONIC GONADOTROPIN [MODELED WITH DIANTENNARY GLYCAN AT ASN78] | | Descriptor: | CHORIONIC GONADOTROPIN, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Erbel, P.J.A, Karimi-Nejad, Y, van Kuik, J.A, Boelens, R, Kamerling, J.P, Vliegenthart, J.F.G. | | Deposit date: | 2000-11-07 | | Release date: | 2000-12-07 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Effects of the N-Linked Glycans on the 3D Structure of the Free A-Subunit of Human Chorionic Gonadotropin
Biochemistry, 39, 2000
|
|
2Y4W
 
 | | Solution structure of human ubiquitin conjugating enzyme Rad6b | | Descriptor: | UBIQUITIN-CONJUGATING ENZYME E2 B | | Authors: | Huang, A, Hibbert, R.G, deJong, R.N, Das, D, Sixma, T.K, Boelens, R. | | Deposit date: | 2011-01-11 | | Release date: | 2011-05-11 | | Last modified: | 2012-06-27 | | Method: | SOLUTION NMR | | Cite: | Symmetry and Asymmetry of the Ring-Ring Dimer of Rad18
J.Mol.Biol., 410, 2011
|
|
1Z60
 
 | | Solution structure of the carboxy-terminal domain of human TFIIH P44 subunit | | Descriptor: | TFIIH basal transcription factor complex p44 subunit, ZINC ION | | Authors: | Kellenberger, E, Dominguez, C, Fribourg, S, Wasielewski, E, Moras, D, Poterszman, A, Boelens, R, Kieffer, B. | | Deposit date: | 2005-03-21 | | Release date: | 2005-04-12 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of TFIIH P44 subunit reveals a novel type of C4C4 ring domain involved in protein-protein interactions.
J.Biol.Chem., 280, 2005
|
|
2KEK
 
 | | Solution structure of a dimer of LAC repressor DNA-binding domain complexed to its natural operator O3 | | Descriptor: | DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*GP*AP*GP*CP*GP*CP*AP*AP*CP*GP*CP*AP*AP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*TP*TP*GP*CP*GP*TP*TP*GP*CP*GP*CP*TP*CP*AP*CP*TP*GP*CP*CP*G)-3'), Lactose operon repressor | | Authors: | Romanuka, J, Folkers, G, Biris, N, Tishchenko, E, Wienk, H, Kaptein, R, Boelens, R. | | Deposit date: | 2009-01-30 | | Release date: | 2009-05-19 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Specificity and affinity of Lac repressor for the auxiliary operators O2 and O3 are explained by the structures of their protein-DNA complexes.
J.Mol.Biol., 390, 2009
|
|
2KEI
 
 | | Refined Solution Structure of a Dimer of LAC repressor DNA-Binding domain complexed to its natural operator O1 | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*GP*AP*TP*AP*AP*CP*AP*AP*TP*TP*T)-3'), DNA (5'-D(P*AP*AP*AP*TP*TP*GP*TP*TP*AP*TP*CP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*TP*C)-3'), Lactose operon repressor | | Authors: | Romanuka, J, Folkers, G, Biris, N, Tishchenko, E, Wienk, H, Kaptein, R, Boelens, R. | | Deposit date: | 2009-01-30 | | Release date: | 2009-05-19 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Specificity and affinity of Lac repressor for the auxiliary operators O2 and O3 are explained by the structures of their protein-DNA complexes.
J.Mol.Biol., 390, 2009
|
|
2KEJ
 
 | | Solution structure of a dimer of LAC repressor DNA-binding domain complexed to its natural operator O2 | | Descriptor: | DNA (5'-D(*GP*AP*AP*AP*TP*GP*TP*GP*AP*GP*CP*GP*AP*GP*TP*AP*AP*CP*AP*AP*CP*CP*G)-3'), DNA (5'-D(P*CP*GP*GP*TP*TP*GP*TP*TP*AP*CP*TP*CP*GP*CP*TP*CP*AP*CP*AP*TP*TP*TP*C)-3'), Lactose operon repressor | | Authors: | Romanuka, J, Folkers, G, Biris, N, Tishchenko, E, Wienk, H, Kaptein, R, Boelens, R. | | Deposit date: | 2009-01-30 | | Release date: | 2009-05-19 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Specificity and affinity of Lac repressor for the auxiliary operators O2 and O3 are explained by the structures of their protein-DNA complexes.
J.Mol.Biol., 390, 2009
|
|
2LKC
 
 | | Free B.st IF2-G2 | | Descriptor: | Translation initiation factor IF-2 | | Authors: | Wienk, H, Tishchenko, E, Belardinelli, R, Tomaselli, S, Dongre, R, Spurio, R, Folkers, G.E, Gualerzi, C.O, Boelens, R. | | Deposit date: | 2011-10-10 | | Release date: | 2012-02-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Dynamics of Bacterial Translation Initiation Factor IF2.
J.Biol.Chem., 287, 2012
|
|
2LKD
 
 | | IF2-G2 GDP complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Translation initiation factor IF-2 | | Authors: | Wienk, H, Tishchenko, E, Belardinelli, R, Tomaselli, S, Dongre, R, Spurio, R, Folkers, G.E, Gualerzi, C.O, Boelens, R. | | Deposit date: | 2011-10-10 | | Release date: | 2012-02-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Dynamics of Bacterial Translation Initiation Factor IF2.
J.Biol.Chem., 287, 2012
|
|
2MH3
 
 | | NMR structure of the basic helix-loop-helix region of the transcriptional repressor HES-1 | | Descriptor: | Transcription factor HES-1 | | Authors: | Wienk, H, Popovic, M, Coglievina, M, Boelens, R, Pongor, S, Pintar, A. | | Deposit date: | 2013-11-15 | | Release date: | 2014-02-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The basic helix-loop-helix region of the transcriptional repressor hairy and enhancer of split 1 is preorganized to bind DNA.
Proteins, 82, 2014
|
|
2N01
 
 | | NMR structure of VirB9 C-terminal domain in complex with VirB7 N-terminal domain from Xanthomonas citri's T4SS | | Descriptor: | VirB7 protein, VirB9 protein | | Authors: | Oliveira, L.C, Souza, D.P, Salinas, R.K, Wienk, H, Boelens, R, Farah, S.C. | | Deposit date: | 2015-03-03 | | Release date: | 2015-09-09 | | Last modified: | 2016-10-19 | | Method: | SOLUTION NMR | | Cite: | VirB7 and VirB9 Interactions Are Required for the Assembly and Antibacterial Activity of a Type IV Secretion System.
Structure, 24, 2016
|
|
2JPD
 
 | | Solution structure of the ERCC1 central domain | | Descriptor: | DNA excision repair protein ERCC-1 | | Authors: | Tripsianes, K, Folkers, G, Zheng, C, Das, D, Grinstead, J.S, Kaptein, R, Boelens, R. | | Deposit date: | 2007-05-06 | | Release date: | 2007-09-04 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Analysis of the XPA and ssDNA-binding surfaces on the central domain of human ERCC1 reveals evidence for subfunctionalization
Nucleic Acids Res., 35, 2007
|
|
2KN7
 
 | | Structure of the XPF-single strand DNA complex | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*GP*GP*CP*TP*GP*A)-3'), DNA repair endonuclease XPF | | Authors: | Das, D, Folkers, G.E, van Dijk, M, Jaspers, N.G.J, Hoeijmakers, J.H.J, Kaptein, R, Boelens, R. | | Deposit date: | 2009-08-16 | | Release date: | 2010-08-04 | | Last modified: | 2012-07-04 | | Method: | SOLUTION NMR | | Cite: | The structure of the XPF-ssDNA complex underscores the distinct roles of the XPF and ERCC1 helix- hairpin-helix domains in ss/ds DNA recognition
Structure, 20, 2012
|
|
2K27
 
 | | Solution structure of Human Pax8 Paired Box Domain | | Descriptor: | Paired box protein Pax-8 | | Authors: | Codutti, L, Esposito, G, Corazza, A, Fogolari, F, Tell, G, Vascotto, C, van Ingen, H, Boelens, R, Viglino, P, Quadrifoglio, F. | | Deposit date: | 2008-03-26 | | Release date: | 2008-09-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of DNA-free Pax-8 Paired Box Domain Accounts for Redox Regulation of Transcriptional Activity in the Pax Protein Family.
J.Biol.Chem., 283, 2008
|
|
2K16
 
 | | Solution structure of the free TAF3 PHD domain | | Descriptor: | Transcription initiation factor TFIID subunit 3, ZINC ION | | Authors: | van Ingen, H, van Schaik, F.M.A, Wienk, H, Timmers, M, Boelens, R. | | Deposit date: | 2008-02-22 | | Release date: | 2008-07-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Recognition of the H3K4me3 mark by the TAF3-PHD finger
To be Published
|
|
2K4D
 
 | | E2-c-Cbl recognition is necessary but not sufficient for ubiquitination activity | | Descriptor: | E3 ubiquitin-protein ligase CBL, ZINC ION | | Authors: | Huang, A, De Jong, R.N, Wienk, H, Winkler, S.G, Timmers, H.T.M, Boelens, R. | | Deposit date: | 2008-06-06 | | Release date: | 2009-01-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | E2-c-Cbl recognition is necessary but not sufficient for ubiquitination activity
J.Mol.Biol., 385, 2009
|
|
2K17
 
 | | Solution structure of the TAF3 PHD domain in complex with a H3K4me3 peptide | | Descriptor: | H3K4me3 peptide, Transcription initiation factor TFIID subunit 3, ZINC ION | | Authors: | van Ingen, H, van Schaik, F.M.A, Wienk, H, Timmers, M, Boelens, R. | | Deposit date: | 2008-02-22 | | Release date: | 2008-07-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Recognition of the H3K4me3 mark by the TAF3-PHD finger
To be Published
|
|
2KXI
 
 | | Solution NMR structure of the apoform of NarE (NMB1343) | | Descriptor: | Uncharacterized protein | | Authors: | Koehler, C, Carlier, L, Veggi, D, Soriani, M, Pizza, M, Boelens, R, Bonvin, A.M.J.J. | | Deposit date: | 2010-05-06 | | Release date: | 2011-03-02 | | Last modified: | 2018-08-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the apoform of NarE (NMB1343)
To be Published
|
|
6YN1
 
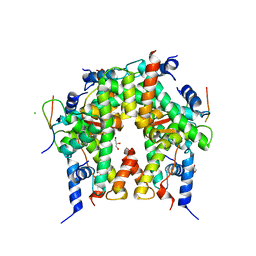 | | Crystal structure of histone chaperone APLF acidic domain bound to the histone H2A-H2B-H3-H4 octamer | | Descriptor: | Aprataxin and PNK-like factor, CHLORIDE ION, GLYCEROL, ... | | Authors: | Corbeski, I, Guo, X, Van Ingen, H, Sixma, T.K. | | Deposit date: | 2020-04-10 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chaperoning of the histone octamer by the acidic domain of DNA repair factor APLF.
Sci Adv, 8, 2022
|
|
