1MB4
 
 | | Crystal structure of aspartate semialdehyde dehydrogenase from vibrio cholerae with NADP and S-methyl-l-cysteine sulfoxide | | Descriptor: | Aspartate-Semialdehyde Dehydrogenase, CYSTEINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Blanco, J, Moore, R.A, Kabaleeswaran, V, Viola, R.E. | | Deposit date: | 2002-08-02 | | Release date: | 2003-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A structural Basis for the Mechanism of Aspartate-beta-semialdehyde Dehydrogenase from Vibrio Cholerae
Protein Sci., 12, 2003
|
|
1MC4
 
 | |
1NWC
 
 | |
1OZA
 
 | |
1NWH
 
 | |
1PQU
 
 | | Crystal Structure of the H277N Mutant of Aspartate Semialdehyde Dehydrogenase from Haemophilus influenzae Bound with NADP, S-methyl cysteine sulfoxide and cacodylate | | Descriptor: | Aspartate-semialdehyde dehydrogenase, CACODYLATE ION, CYSTEINE, ... | | Authors: | Blanco, J, Moore, R.A, Viola, R.E. | | Deposit date: | 2003-06-19 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PS8
 
 | | Crystal Structure of the R270K Mutant of Aspartate Semialdehyde dehydrogenase from Haemophilus influenzae | | Descriptor: | Aspartate semialdehyde dehydrogenase | | Authors: | Blanco, J, Moore, R.A, Faehnle, C.R, Coe, D.M, Viola, R.E. | | Deposit date: | 2003-06-20 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PU2
 
 | | Crystal Structure of the K246R Mutant of Aspartate Semialdehyde Dehydrogenase from Haemophilus influenzae | | Descriptor: | Aspartate-semialdehyde dehydrogenase | | Authors: | Blanco, J, Moore, R.A, Faehnle, C.R, Coe, D.M, Viola, R.E. | | Deposit date: | 2003-06-23 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PQP
 
 | | Crystal Structure of the C136S Mutant of Aspartate Semialdehyde Dehydrogenase from Haemophilus influenzae Bound with Aspartate Semialdehyde and Phosphate | | Descriptor: | Aspartate-semialdehyde dehydrogenase, L-HOMOSERINE, PHOSPHATE ION | | Authors: | Blanco, J, Moore, R.A, Faehnle, C.R, Viola, R.E. | | Deposit date: | 2003-06-18 | | Release date: | 2004-08-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1Q2X
 
 | | Crystal Structure of the E243D Mutant of Aspartate Semialdehyde Dehydrogenase from Haemophilus influenzae bound with substrate aspartate semialdehyde | | Descriptor: | Aspartate-semialdehyde dehydrogenase | | Authors: | Blanco, J, Moore, R.A, Faehnle, C.R, Coe, D.M, Viola, R.E. | | Deposit date: | 2003-07-26 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1NX6
 
 | |
1PR3
 
 | | Crystal Structure of the R103K Mutant of Aspartate Semialdehyde dehydrogenase from Haemophilus influenzae | | Descriptor: | Aspartate semialdehyde dehydrogenase, PHOSPHATE ION | | Authors: | Blanco, J, Moore, R.A, Faehnle, C.R, Coe, D.M, Viola, R.E. | | Deposit date: | 2003-06-19 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4P7E
 
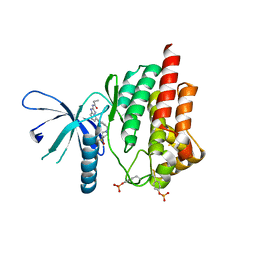 | | Triazolopyridine compounds as selective JAK1 inhibitors: from hit identification to GLPG0634 | | Descriptor: | N-(5-{4-[(1,1-dioxidothiomorpholin-4-yl)methyl]phenyl}[1,2,4]triazolo[1,5-a]pyridin-2-yl)cyclopropanecarboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Menet, C.C.J, Fletcher, S, Van Lommen, G, Geney, R, Blanc, J, Smits, K, Jouannigot, N, van der Aar, E.M, Clement-Lacroix, P, Lepescheux, L, Galien, R, Vayssiere, B, Nelles, L, Christophe, T, Brys, R, Uhring, M, Ciesielski, F, Van Rompaey, L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Triazolopyridines as Selective JAK1 Inhibitors: From Hit Identification to GLPG0634.
J.Med.Chem., 57, 2014
|
|
4ZJH
 
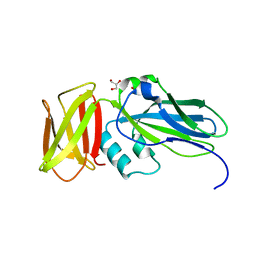 | | Crystal structure of native alpha-2-macroglobulin from Escherichia coli spanning domains NIE-MG1. | | Descriptor: | ACETATE ION, GLYCEROL, alpha-2-Macroglobulin | | Authors: | Garcia-Ferrer, I, Arede, P, Gomez-Blanco, J, Luque, D, Duquerroy, S, Caston, J.R, Goulas, T, Gomis-Ruth, X.F. | | Deposit date: | 2015-04-29 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional insights into Escherichia coli alpha 2-macroglobulin endopeptidase snap-trap inhibition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A42
 
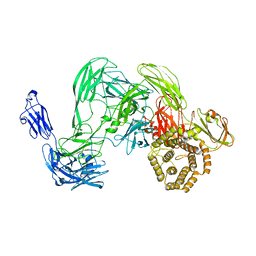 | | Cryo-EM single particle 3D reconstruction of the native conformation of E. coli alpha-2-macroglobulin (ECAM) | | Descriptor: | UNCHARACTERIZED LIPOPROTEIN YFHM | | Authors: | Garcia-Ferrer, I, Arede, P, Gomez-Blanco, J, Luque, D, Duquerroy, S, Caston, J.R, Goulas, T, Gomis-Ruth, F.X. | | Deposit date: | 2015-06-04 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Structural and Functional Insights Into Escherichia Coli Alpha2- Macroglobulin Endopeptidase Snap-Trap Inhibition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2AM9
 
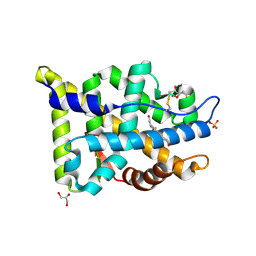 | | Crystal structure of human androgen receptor ligand binding domain in complex with testosterone | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Androgen receptor, GLYCEROL, ... | | Authors: | Pereira de Jesus-Tran, K, Cote, P.-L, Cantin, L, Blanchet, J, Labrie, F, Breton, R. | | Deposit date: | 2005-08-09 | | Release date: | 2006-05-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Comparison of crystal structures of human androgen receptor ligand-binding domain complexed with various agonists reveals molecular determinants responsible for binding affinity.
Protein Sci., 15, 2006
|
|
2AMA
 
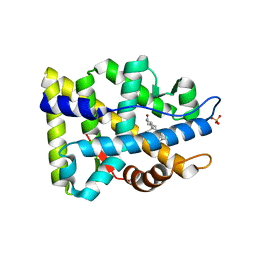 | | Crystal structure of human androgen receptor ligand binding domain in complex with dihydrotestosterone | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, SULFATE ION | | Authors: | Pereira de Jesus-Tran, K, Cote, P.-L, Cantin, L, Blanchet, J, Labrie, F, Breton, R. | | Deposit date: | 2005-08-09 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparison of crystal structures of human androgen receptor ligand-binding domain complexed with various agonists reveals molecular determinants responsible for binding affinity.
Protein Sci., 15, 2006
|
|
6QI5
 
 | | Near Atomic Structure of an Atadenovirus Shows a possible gene duplication event and Intergenera Variations in Cementing Proteins | | Descriptor: | Hexon protein, PIIIa, Penton protein, ... | | Authors: | Condezo, G.N, Marabini, R, Gomez-Blanco, J, SanMartin, C. | | Deposit date: | 2019-01-17 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Near-atomic structure of an atadenovirus reveals a conserved capsid-binding motif and intergenera variations in cementing proteins.
Sci Adv, 7, 2021
|
|
6W7N
 
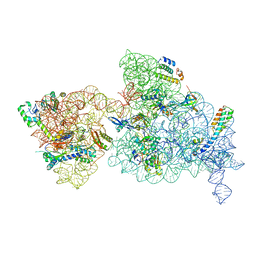 | | 30S-Inactive-low-Mg2+ Class A | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S12, ... | | Authors: | Jahagirdar, D, Jha, V, Basu, B, Gomez-Blanco, J, Vargas, J, Ortega, J. | | Deposit date: | 2020-03-19 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Alternative conformations and motions adopted by 30S ribosomal subunits visualized by cryo-electron microscopy.
Rna, 26, 2020
|
|
6W7W
 
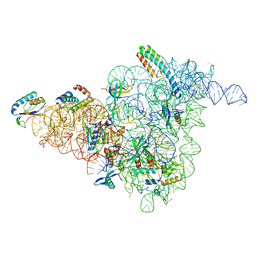 | | 30S-Inactive-low-Mg2+ Class B | | Descriptor: | 16S rRNA, 30S ribosomal protein S12, 30S ribosomal protein S15, ... | | Authors: | Jahagirdar, D, Jha, V, Basu, B, Gomez-Blanco, J, Vargas, J, Ortega, J. | | Deposit date: | 2020-03-19 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Alternative conformations and motions adopted by 30S ribosomal subunits visualized by cryo-electron microscopy.
Rna, 26, 2020
|
|
6W6K
 
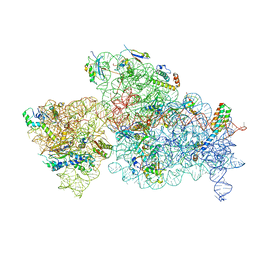 | | 30S-Activated-high-Mg2+ | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Jahagirdar, D, Jha, V, Basu, B, Gomez-Blanco, J, Vargas, J, Ortega, J. | | Deposit date: | 2020-03-17 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Alternative conformations and motions adopted by 30S ribosomal subunits visualized by cryo-electron microscopy.
Rna, 26, 2020
|
|
6W77
 
 | | 30S-Inactivated-high-Mg2+ Class A | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Jahagirdar, D, Jha, V, Basu, B, Gomez-Blanco, J, Vargas, J, Ortega, J. | | Deposit date: | 2020-03-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Alternative conformations and motions adopted by 30S ribosomal subunits visualized by cryo-electron microscopy.
Rna, 26, 2020
|
|
6W7M
 
 | | 30S-Inactive-high-Mg2+ + carbon layer | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Jahagirdar, D, Jha, V, Basu, B, Gomez-Blanco, J, Vargas, J, Ortega, J. | | Deposit date: | 2020-03-19 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Alternative conformations and motions adopted by 30S ribosomal subunits visualized by cryo-electron microscopy.
Rna, 26, 2020
|
|
3ZUE
 
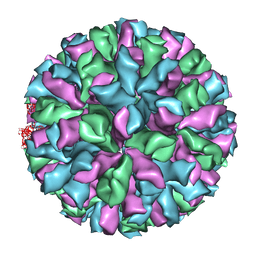 | | Rabbit Hemorrhagic Disease Virus (RHDV)capsid protein | | Descriptor: | CAPSID STRUCTURAL PROTEIN VP60 | | Authors: | Luque, D, Gonzalez, J.M, Gomez-Blanco, J, Marabini, R, Chichon, J, Mena, I, Angulo, I, Carrascosa, J.L, Verdaguer, N, Trus, B.L, Barcena, J, Caston, J.R. | | Deposit date: | 2011-07-18 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.3 Å) | | Cite: | Epitope Insertion at the N-Terminal Molecular Switch of the Rabbit Hemorrhagic Disease Virus T=3 Capsid Protein Leads to Larger T=4 Capsids.
J.Virol., 86, 2012
|
|
8C89
 
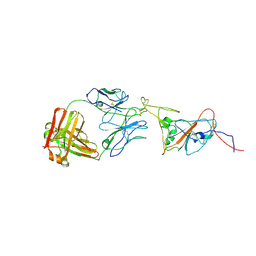 | | SARS-CoV-2 spike in complex with the 17T2 neutralizing antibody Fab fragment (local refinement of RBD and Fab) | | Descriptor: | 17T2 Fab heavy chain, 17T2 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Modrego, A, Carlero, D, Bueno-Carrasco, M.T, Santiago, C, Carolis, C, Arranz, R, Blanco, J, Magri, G. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.41 Å) | | Cite: | A monoclonal antibody targeting a large surface of the receptor binding motif shows pan-neutralizing SARS-CoV-2 activity.
Nat Commun, 15, 2024
|
|
