2YDL
 
 | | Crystal structure of SH3C from CIN85 | | Descriptor: | SH3 DOMAIN-CONTAINING KINASE-BINDING PROTEIN 1 | | Authors: | Bravo, J, Cardenes, N. | | Deposit date: | 2011-03-22 | | Release date: | 2012-03-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Distinct Ubiquitin Binding Modes Exhibited by SH3 Domains: Molecular Determinants and Functional Implications.
Plos One, 8, 2013
|
|
7PON
 
 | |
7PNO
 
 | | C terminal domain of Nipah Virus Phosphoprotein fused to the Ntail alpha more of the Nucleoprotein. | | Descriptor: | Phosphoprotein, alpha MoRE of Nipah virus Nucleoprotein tail | | Authors: | Bourhis, J.M, Yabukaski, F, Tarbouriech, N, Jamin, M. | | Deposit date: | 2021-09-07 | | Release date: | 2022-04-20 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Dynamics of the C-terminal X Domain of Nipah and Hendra Viruses Controls the Attachment to the C-terminal Tail of the Nucleocapsid Protein.
J.Mol.Biol., 434, 2022
|
|
4HEO
 
 | |
4RMW
 
 | |
4RMU
 
 | |
4RMV
 
 | |
6YS3
 
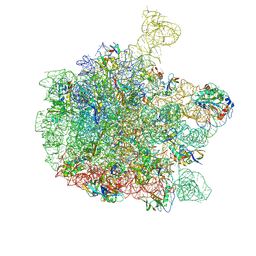 | | Cryo-EM structure of the 50S ribosomal subunit at 2.58 Angstroms with modeled GBC SecM peptide | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Schulte, L, Reitz, J, Kudlinzki, D, Hodirnau, V.V, Frangakis, A, Schwalbe, H. | | Deposit date: | 2020-04-20 | | Release date: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Cryo-EM structure of the 50S ribosomal subunit at 2.58 Angstroms with modeled GBC SecM peptide
Nat Commun, 2020
|
|
6QDW
 
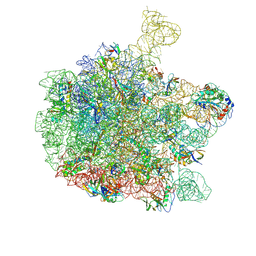 | | Cryo-EM structure of the 50S ribosomal subunit at 2.83 Angstroms with modeled GBC SecM peptide | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Schulte, L, Reitz, J, Hodirnau, V.V, Kudlinzki, D, Mao, J, Glaubitz, C, Frangakis, A, Schwalbe, H. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-15 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cysteine oxidation and disulfide formation in the ribosomal exit tunnel.
Nat Commun, 11, 2020
|
|
5CSG
 
 | |
5CSB
 
 | | The crystal structure of beta2-microglobulin D76N mutant at room temperature | | Descriptor: | Beta-2-microglobulin | | Authors: | de Rosa, M, Mota, C.S, de Sanctis, D, Bolognesi, M, Ricagno, S. | | Deposit date: | 2015-07-23 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | Conformational dynamics in crystals reveal the molecular bases for D76N beta-2 microglobulin aggregation propensity.
Nat Commun, 9, 2018
|
|
5CS7
 
 | | The crystal structure of wt beta2-microglobulin at room temperature | | Descriptor: | Beta-2-microglobulin | | Authors: | de Rosa, M, Mota, C.S, de Sanctis, D, Bolognesi, M, Ricagno, S. | | Deposit date: | 2015-07-23 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational dynamics in crystals reveal the molecular bases for D76N beta-2 microglobulin aggregation propensity.
Nat Commun, 9, 2018
|
|
5A7L
 
 | |
3ZVU
 
 | | Structure of the PYR1 His60Pro mutant in complex with the HAB1 phosphatase and Abscisic acid | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABSCISIC ACID RECEPTOR PYR1, MANGANESE (II) ION, ... | | Authors: | Betz, K, Dupeux, F, Santiago, J, Rodriguez, P.L, Marquez, J.A. | | Deposit date: | 2011-07-27 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Thermodynamic Switch Modulates Abscisic Acid Receptor Sensitivity.
Embo J., 30, 2011
|
|
4H2D
 
 | | Crystal structure of NDOR1 | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH-dependent diflavin oxidoreductase 1 | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mikolajczyk, M, Jaiswal, D, Winkelmann, J. | | Deposit date: | 2012-09-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular view of an electron transfer process essential for iron-sulfur protein biogenesis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1M9L
 
 | | Relaxation-based Refined Structure Of Chlamydomonas Outer Arm Dynein Light Chain 1 | | Descriptor: | Outer Arm Dynein Light Chain 1 | | Authors: | Wu, H.W, Maciejewski, M.W, Marintchev, A, Benashski, S.E, Mullen, G.P, King, S.M. | | Deposit date: | 2002-07-29 | | Release date: | 2003-03-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Relaxation-based structure refinement and backbone molecular dynamics of the
Dynein motor domain-associated light chain
Biochemistry, 42, 2003
|
|
3PMK
 
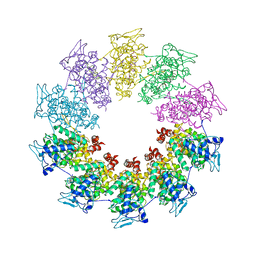 | | Crystal structure of the Vesicular Stomatitis Virus RNA free nucleoprotein/phosphoprotein complex | | Descriptor: | Nucleocapsid protein, Phosphoprotein | | Authors: | Leyrat, C, Yabukarski, F, Tarbouriech, N, Ruigrok, R.W.H, Jamin, M. | | Deposit date: | 2010-11-17 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structure of the Vesicular Stomatitis Virus N0-P Complex
Plos Pathog., 7, 2011
|
|
2KRN
 
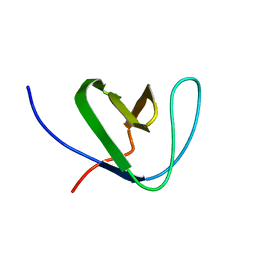 | |
1O15
 
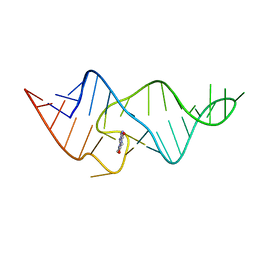 | |
3ZBE
 
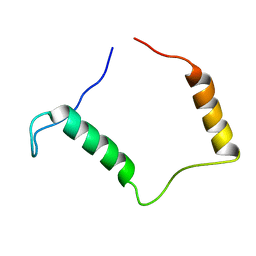 | |
7NYK
 
 | |
7NYM
 
 | | Mutant V517A - SH3 domain of JNK-interacting Protein 1 (JIP1) | | Descriptor: | HEXAETHYLENE GLYCOL, PHOSPHATE ION, SH3 domain of JNK-interacting Protein 1 (JIP1), ... | | Authors: | Perez, L.M, Ielasi, F.S, Palencia, A, Jensen, M.R. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.614 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
7NYN
 
 | | Mutant Y526A of SH3 domain of JNK-interacting Protein 1 (JIP1) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Perez, L.M, Ielasi, F.S, Palencia, A, Jensen, M.R. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.537 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
7NYL
 
 | | Mutant H493A of SH3 domain of JNK-interacting Protein 1 (JIP1) | | Descriptor: | SH3 domain of JNK-interacting Protein 1 (JIP1), TETRAETHYLENE GLYCOL, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Perez, L.M, Ielasi, F.S, Palencia, A, Jensen, M.R. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
7NYO
 
 | | Mutant A541L of SH3 domain of JNK-interacting Protein 1 (JIP1) | | Descriptor: | 1,2-ETHANEDIOL, SH3 domain of JNK-interacting Protein 1 (JIP1), SULFATE ION, ... | | Authors: | Perez, L.M, Ielasi, F.S, Palencia, A, Jensen, M.R. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
