1QVP
 
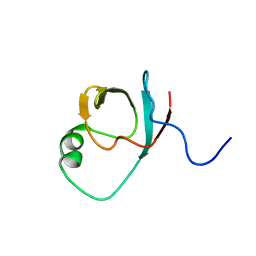 | | C terminal SH3-like domain from Diphtheria toxin Repressor residues 144-226. | | Descriptor: | Diphtheria toxin repressor | | Authors: | Wylie, G.P, Rangachari, V, Bienkiewicz, E.A, Marin, V, Bhattacharya, N, Love, J.F, Murphy, J.R, Logan, T.M. | | Deposit date: | 2003-08-28 | | Release date: | 2004-11-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Prolylpeptide binding by the prokaryotic SH3-like domain of the diphtheria toxin repressor: a regulatory switch.
Biochemistry, 44, 2005
|
|
7MT0
 
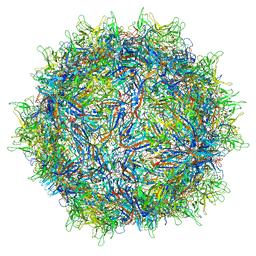 | | Structure of the adeno-associated virus 9 capsid at pH 7.4 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-12 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MTZ
 
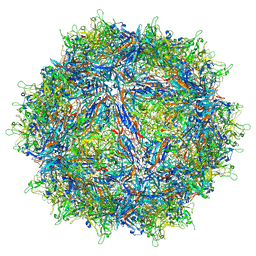 | | Structure of the adeno-associated virus 9 capsid at pH pH 7.4 in complex with terminal galactose | | Descriptor: | Capsid protein VP1, beta-D-galactopyranose | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-14 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
6WH3
 
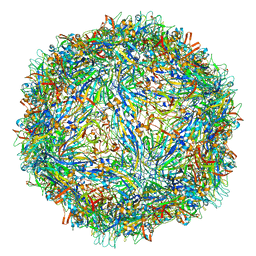 | | Capsid structure of Penaeus monodon metallodensovirus at pH 8.2 | | Descriptor: | CALCIUM ION, Penaeus monodon metallodensovirus major capsid protein | | Authors: | Penzes, J.J, Pham, H.T, Chipman, P, Bhattacharya, N, McKenna, R, Agbandje-McKenna, M, Tijssen, P. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Molecular biology and structure of a novel penaeid shrimp densovirus elucidate convergent parvoviral host capsid evolution.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7MTG
 
 | | Structure of the adeno-associated virus 9 capsid at pH 6.0 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MTP
 
 | | Structure of the adeno-associated virus 9 capsid at pH 5.5 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MTW
 
 | | Structure of the adeno-associated virus 9 capsid at pH 4.0 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MUA
 
 | | Structure of the adeno-associated virus 9 capsid at pH pH 5.5 in complex with terminal galactose | | Descriptor: | Capsid protein VP1, beta-D-galactopyranose | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-14 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
6WH7
 
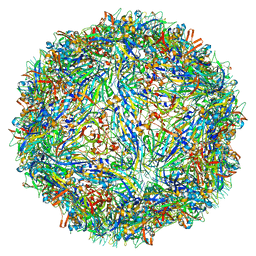 | | Capsid structure of Penaeus monodon metallodensovirus following EDTA treatment | | Descriptor: | Penaeus monodon metallodensovirus major capsid protein | | Authors: | Penzes, J.J, Pham, H.T, Chipman, P, Bhattacharya, N, McKenna, R, Agbandje-McKenna, M, Tijssen, P. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Molecular biology and structure of a novel penaeid shrimp densovirus elucidate convergent parvoviral host capsid evolution.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1QW1
 
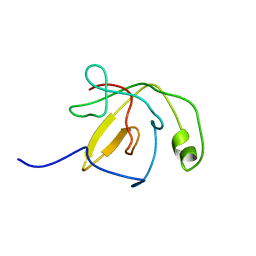 | | Solution Structure of the C-Terminal Domain of DtxR residues 110-226 | | Descriptor: | Diphtheria toxin repressor | | Authors: | Wylie, G.P, Rangachari, V, Bienkiewicz, E.A, Love, J.F, Murphy, J.R, Logan, T.M. | | Deposit date: | 2003-08-29 | | Release date: | 2005-03-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Prolylpeptide binding by the prokaryotic SH3-like domain of the diphtheria toxin repressor: a regulatory switch.
Biochemistry, 44, 2005
|
|
4RDP
 
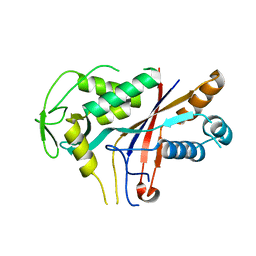 | | Crystal structure of Cmr4 | | Descriptor: | CRISPR system Cmr subunit Cmr4 | | Authors: | Shao, Y, Tang, L, Li, H. | | Deposit date: | 2014-09-19 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Essential Structural and Functional Roles of the Cmr4 Subunit in RNA Cleavage by the Cmr CRISPR-Cas Complex.
Cell Rep, 9, 2014
|
|
6NXE
 
 | |
6O9R
 
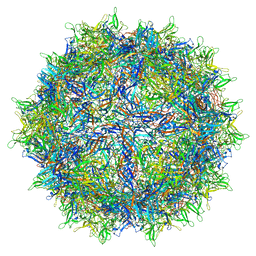 | |
7L6B
 
 | | The empty AAV12 capsid | | Descriptor: | VP1 | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2020-12-23 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Completion of the AAV Structural Atlas: Serotype Capsid Structures Reveals Clade-Specific Features.
Viruses, 13, 2021
|
|
7L6I
 
 | | The empty AAV13 capsid | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2020-12-23 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Completion of the AAV Structural Atlas: Serotype Capsid Structures Reveals Clade-Specific Features.
Viruses, 13, 2021
|
|
7L5Q
 
 | | The empty AAV7 capsid | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2020-12-22 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Completion of the AAV Structural Atlas: Serotype Capsid Structures Reveals Clade-Specific Features.
Viruses, 13, 2021
|
|
7L6H
 
 | | The genome-containing AAV13 capsid | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Capsid protein | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2020-12-23 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Completion of the AAV Structural Atlas: Serotype Capsid Structures Reveals Clade-Specific Features.
Viruses, 13, 2021
|
|
7L6A
 
 | | The genome-containing AAV12 capsid | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, VP1 | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2020-12-23 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Completion of the AAV Structural Atlas: Serotype Capsid Structures Reveals Clade-Specific Features.
Viruses, 13, 2021
|
|
7L5U
 
 | | The full AAV7 capsid | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Capsid protein | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2020-12-22 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Completion of the AAV Structural Atlas: Serotype Capsid Structures Reveals Clade-Specific Features.
Viruses, 13, 2021
|
|
7L6E
 
 | | The genome-containing AAV11 capsid | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Capsid protein | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2020-12-23 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Completion of the AAV Structural Atlas: Serotype Capsid Structures Reveals Clade-Specific Features.
Viruses, 13, 2021
|
|
7L6F
 
 | | The empty AAV11 capsid | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2020-12-23 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Completion of the AAV Structural Atlas: Serotype Capsid Structures Reveals Clade-Specific Features.
Viruses, 13, 2021
|
|
7U97
 
 | | SAAV pH 4.0 capsid structure | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2022-03-10 | | Release date: | 2022-04-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Characterization of the Serpentine Adeno-Associated Virus (SAAV) Capsid Structure: Receptor Interactions and Antigenicity.
J.Virol., 96, 2022
|
|
7U95
 
 | | SAAV pH 6.0 capsid structure | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2022-03-10 | | Release date: | 2022-04-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Characterization of the Serpentine Adeno-Associated Virus (SAAV) Capsid Structure: Receptor Interactions and Antigenicity.
J.Virol., 96, 2022
|
|
7U96
 
 | | SAAV pH 5.5 capsid structure | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2022-03-10 | | Release date: | 2022-04-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Characterization of the Serpentine Adeno-Associated Virus (SAAV) Capsid Structure: Receptor Interactions and Antigenicity.
J.Virol., 96, 2022
|
|
7U94
 
 | | SAAV pH 7.4 capsid structure | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2022-03-10 | | Release date: | 2022-04-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Characterization of the Serpentine Adeno-Associated Virus (SAAV) Capsid Structure: Receptor Interactions and Antigenicity.
J.Virol., 96, 2022
|
|
