1R18
 
 | | Drosophila protein isoaspartyl methyltransferase with S-adenosyl-L-homocysteine | | Descriptor: | Protein-L-isoaspartate(D-aspartate)-O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Bennett, E.J, Bjerregaard, J, Knapp, J.E, Chavous, D.A, Friedman, A.M, Royer Jr, W.E, O'Connor, C.M. | | Deposit date: | 2003-09-23 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic implications from the Drosophila protein L-isoaspartyl methyltransferase structure and site-directed mutagenesis.
Biochemistry, 42, 2003
|
|
4GBA
 
 | | DCNL complex with N-terminally acetylated NEDD8 E2 peptide | | Descriptor: | DCN1-like protein 3, NEDD8-conjugating enzyme UBE2F | | Authors: | Monda, J.K, Scott, D.C, Miller, D.J, Harper, J.W, Bennett, E.J, Schulman, B.A. | | Deposit date: | 2012-07-26 | | Release date: | 2012-11-28 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Conservation of Distinctive N-terminal Acetylation-Dependent Interactions across a Family of Mammalian NEDD8 Ligation Enzymes.
Structure, 21, 2013
|
|
4GAO
 
 | | DCNL complex with N-terminally acetylated NEDD8 E2 peptide | | Descriptor: | BROMIDE ION, DCN1-like protein 2, NEDD8-conjugating enzyme Ubc12 | | Authors: | Monda, J.K, Scott, D.C, Miller, D.J, Harper, J.W, Bennett, E.J, Schulman, B.A. | | Deposit date: | 2012-07-25 | | Release date: | 2012-11-28 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structural Conservation of Distinctive N-terminal Acetylation-Dependent Interactions across a Family of Mammalian NEDD8 Ligation Enzymes.
Structure, 21, 2013
|
|
3TDI
 
 | | yeast Cul1WHB-Dcn1P acetylated Ubc12N complex | | Descriptor: | Defective in cullin neddylation protein 1, NEDD8-conjugating enzyme UBC12 | | Authors: | Scott, D.C, Monda, J.K, Bennett, E.J, Harper, J.W, Schulman, B.A. | | Deposit date: | 2011-08-11 | | Release date: | 2011-10-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex.
Science, 334, 2011
|
|
3TDZ
 
 | | N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex: Structure of a human Cul1WHB-Dcn1P-stapled acetylated Ubc12N complex | | Descriptor: | Cullin-1, DCN1-like protein 1, STAPLED PEPTIDE | | Authors: | Scott, D.C, Monda, J.K, Bennett, E.J, Harper, J.W, Schulman, B.A. | | Deposit date: | 2011-08-11 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex.
Science, 334, 2011
|
|
3TDU
 
 | | N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex: Structure of a human Cul1WHB-Dcn1P-acetylated Ubc12N complex | | Descriptor: | Cullin-1, DCN1-like protein 1, NEDD8-conjugating enzyme Ubc12 | | Authors: | Scott, D.C, Monda, J.K, Bennett, E.J, Harper, J.W, Schulman, B.A. | | Deposit date: | 2011-08-11 | | Release date: | 2011-10-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex.
Science, 334, 2011
|
|
6ZVI
 
 | | Mbf1-ribosome complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S0-A, 40S ribosomal protein S10-A, ... | | Authors: | Best, K.M, Denk, T, Cheng, J, Thoms, M, Berninghausen, O, Beckmann, R. | | Deposit date: | 2020-07-24 | | Release date: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | EDF1 coordinates cellular responses to ribosome collisions.
Elife, 9, 2020
|
|
6ZVH
 
 | | EDF1-ribosome complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Best, K.M, Denk, T, Cheng, J, Thoms, M, Berninghausen, O, Beckmann, R. | | Deposit date: | 2020-07-24 | | Release date: | 2020-08-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | EDF1 coordinates cellular responses to ribosome collisions.
Elife, 9, 2020
|
|
7MWD
 
 | | HUWE1 in map with focus on HECT | | Descriptor: | E3 ubiquitin-protein ligase HUWE1 | | Authors: | Hunkeler, M, Fischer, E.S. | | Deposit date: | 2021-05-16 | | Release date: | 2021-07-28 | | Last modified: | 2021-09-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Solenoid architecture of HUWE1 contributes to ligase activity and substrate recognition.
Mol.Cell, 81, 2021
|
|
7MWF
 
 | | HUWE1 in map with focus on interface | | Descriptor: | E3 ubiquitin-protein ligase HUWE1 | | Authors: | Hunkeler, M, Fischer, E.S. | | Deposit date: | 2021-05-16 | | Release date: | 2021-07-28 | | Last modified: | 2021-09-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Solenoid architecture of HUWE1 contributes to ligase activity and substrate recognition.
Mol.Cell, 81, 2021
|
|
7MWE
 
 | | HUWE1 in map with focus on WWE | | Descriptor: | E3 ubiquitin-protein ligase HUWE1 | | Authors: | Hunkeler, M, Fischer, E.S. | | Deposit date: | 2021-05-16 | | Release date: | 2021-07-28 | | Last modified: | 2021-09-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Solenoid architecture of HUWE1 contributes to ligase activity and substrate recognition.
Mol.Cell, 81, 2021
|
|
7MOP
 
 | | Cryo-EM structure of human HUWE1 in complex with DDIT4 | | Descriptor: | DNA damage-inducible transcript 4 protein, E3 ubiquitin-protein ligase HUWE1 | | Authors: | Hunkeler, M, Fischer, E.S. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-28 | | Last modified: | 2021-09-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Solenoid architecture of HUWE1 contributes to ligase activity and substrate recognition.
Mol.Cell, 81, 2021
|
|
7JQ9
 
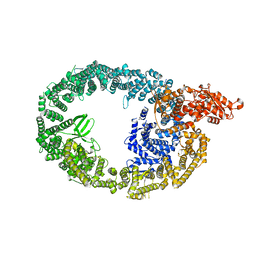 | | Cryo-EM structure of human HUWE1 | | Descriptor: | E3 ubiquitin-protein ligase HUWE1 | | Authors: | Hunkeler, M, Fischer, E.S. | | Deposit date: | 2020-08-10 | | Release date: | 2021-07-28 | | Last modified: | 2021-09-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Solenoid architecture of HUWE1 contributes to ligase activity and substrate recognition.
Mol.Cell, 81, 2021
|
|
