1AF0
 
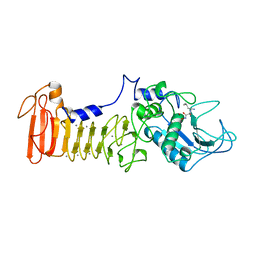 | | SERRATIA PROTEASE IN COMPLEX WITH INHIBITOR | | Descriptor: | CALCIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-hydroxy-L-alaninamide, SERRATIA PROTEASE, ... | | Authors: | Baumann, U. | | Deposit date: | 1997-03-20 | | Release date: | 1998-03-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the 50 kDa Metallo Protease from S. Marcescens in Complex with the Synthetic Inhibitor Cbz-Leu-Ala-Nhoh
To be Published
|
|
1SAT
 
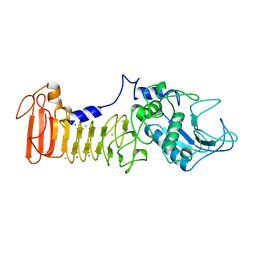 | |
1SMP
 
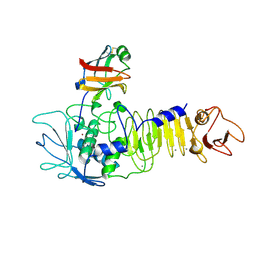 | | CRYSTAL STRUCTURE OF A COMPLEX BETWEEN SERRATIA MARCESCENS METALLO-PROTEASE AND AN INHIBITOR FROM ERWINIA CHRYSANTHEMI | | Descriptor: | CALCIUM ION, ERWINIA CHRYSANTHEMI INHIBITOR, SERRATIA METALLO PROTEINASE, ... | | Authors: | Baumann, U, Bauer, M, Letoffe, S, Delepelaire, P, Wandersman, C. | | Deposit date: | 1995-01-13 | | Release date: | 1996-04-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a complex between Serratia marcescens metallo-protease and an inhibitor from Erwinia chrysanthemi.
J.Mol.Biol., 248, 1995
|
|
1HLE
 
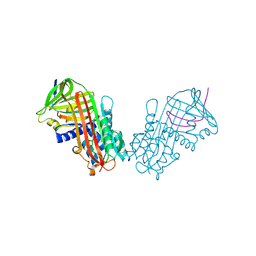 | | CRYSTAL STRUCTURE OF CLEAVED EQUINE LEUCOCYTE ELASTASE INHIBITOR DETERMINED AT 1.95 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, HORSE LEUKOCYTE ELASTASE INHIBITOR | | Authors: | Baumann, U, Bode, W, Huber, R, Travis, J, Potempa, J. | | Deposit date: | 1992-04-13 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of cleaved equine leucocyte elastase inhibitor determined at 1.95 A resolution.
J.Mol.Biol., 226, 1992
|
|
2ACH
 
 | | CRYSTAL STRUCTURE OF CLEAVED HUMAN ALPHA1-ANTICHYMOTRYPSIN AT 2.7 ANGSTROMS RESOLUTION AND ITS COMPARISON WITH OTHER SERPINS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA 1-ANTICHYMOTRYPSIN, PHOSPHATE ION, ... | | Authors: | Baumann, U, Huber, R, Bode, W, Grosse, D, Lesjak, M, Laurell, C.B. | | Deposit date: | 1993-04-26 | | Release date: | 1993-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of cleaved human alpha 1-antichymotrypsin at 2.7 A resolution and its comparison with other serpins.
J.Mol.Biol., 218, 1991
|
|
1KAP
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE ALKALINE PROTEASE OF PSEUDOMONAS AERUGINOSA: A TWO-DOMAIN PROTEIN WITH A CALCIUM BINDING PARALLEL BETA ROLL MOTIF | | Descriptor: | ALKALINE PROTEASE, CALCIUM ION, TETRAPEPTIDE (GLY SER ASN SER), ... | | Authors: | Baumann, U, Wu, S, Flaherty, K.M, Mckay, D.B. | | Deposit date: | 1995-06-08 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Three-dimensional structure of the alkaline protease of Pseudomonas aeruginosa: a two-domain protein with a calcium binding parallel beta roll motif.
EMBO J., 12, 1993
|
|
9G7G
 
 | | Structure of the clippase PaJOS from Pigmentiphaga aceris | | Descriptor: | PaJOS, Polyubiquitin-B, prop-2-en-1-amine | | Authors: | Baumann, U, Uthoff, M, Hermanns, T, Hofmann, K. | | Deposit date: | 2024-07-21 | | Release date: | 2025-04-23 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A family of bacterial Josephin-like deubiquitinases with an irreversible cleavage mode.
Mol.Cell, 85, 2025
|
|
9FN4
 
 | | DUBS Parachlamydia sp. PcJOS | | Descriptor: | L(+)-TARTARIC ACID, Peptidase C39-like domain-containing protein, Ubiquitin | | Authors: | Baumann, U, Hermanns, T. | | Deposit date: | 2024-06-09 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A family of bacterial Josephin-like deubiquitinases with an irreversible cleavage mode.
Mol.Cell, 85, 2025
|
|
9FPA
 
 | |
1GO8
 
 | | The metzincin's methionine: PrtC M226L mutant | | Descriptor: | CALCIUM ION, PHOSPHATE ION, PROTEASE C, ... | | Authors: | Baumann, U. | | Deposit date: | 2001-10-20 | | Release date: | 2002-10-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Complex between Pseudomonas Aeruginosa Alkaline Protease and its Cognate Inhibitor: Inhibition by a Zinc-NH2 Coordinative Bond
J.Biol.Chem., 276, 2001
|
|
1K7Q
 
 | | PrtC from Erwinia chrysanthemi: E189A mutant | | Descriptor: | CALCIUM ION, SECRETED PROTEASE C, ZINC ION | | Authors: | Baumann, U, Hege, T. | | Deposit date: | 2001-10-20 | | Release date: | 2002-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protease C of Erwinia chrysanthemi: The Crystal Structure and Role of Amino Acids Y228 and E189
J.Mol.Biol., 314, 2001
|
|
1K7I
 
 | | PrtC from Erwinia chrysanthemi: Y228F mutant | | Descriptor: | CALCIUM ION, ZINC ION, secreted protease C | | Authors: | Baumann, U, Hege, T. | | Deposit date: | 2001-10-19 | | Release date: | 2002-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Protease C of Erwinia chrysanthemi: The Crystal Structure and Role of Amino Acids Y228 and E189
J.Mol.Biol., 314, 2001
|
|
5D88
 
 | | The Structure of the U32 Peptidase Mk0906 | | Descriptor: | ACETATE ION, Predicted protease of the collagenase family, ZINC ION | | Authors: | Baumann, U, Schacherl, M, Monatda, A.A.M. | | Deposit date: | 2015-08-15 | | Release date: | 2015-12-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The first crystal structure of the peptidase domain of the U32 peptidase family.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1UWE
 
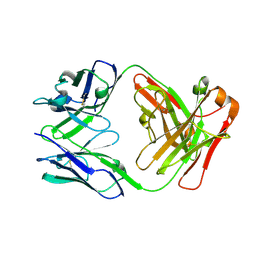 | |
1UWG
 
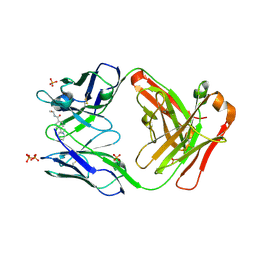 | |
4UWX
 
 | | Structure of liprin-alpha3 in complex with mDia1 Diaphanous- inhibitory domain | | Descriptor: | LIPRIN-ALPHA-3, NICKEL (II) ION, PROTEIN DIAPHANOUS HOMOLOG 1, ... | | Authors: | Brenig, J, de Boor, S, Knyphausen, P, Kuhlmann, N, Wroblowski, S, Baldus, L, Scislowski, L, Artz, O, Trauschies, P, Baumann, U, Neundorf, I, Lammers, M. | | Deposit date: | 2014-08-15 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Basis for the Inhibitory Effect of Liprin-Alpha3 on Mouse Diaphanous 1 (Mdia1) Function.
J.Biol.Chem., 290, 2015
|
|
7BFI
 
 | |
7NDL
 
 | | Crystal structure of human GFAT-1 S205D | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Isoform 2 of Glutamine-fructose-6-phosphate aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Baumann, U, Denzel, M.S. | | Deposit date: | 2021-02-02 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.223 Å) | | Cite: | Protein kinase A controls the hexosamine pathway by tuning the feedback inhibition of GFAT-1.
Nat Commun, 12, 2021
|
|
8CMR
 
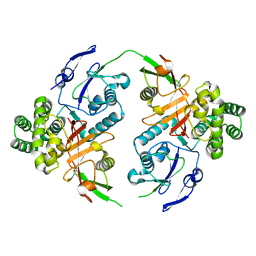 | | Linear specific OTU-type DUB SnOTU from the pathogen S. negenvensis in complex with linear di-ubiquitin | | Descriptor: | OTU domain-containing protein, Polyubiquitin-B | | Authors: | Uthoff, M, Hermanns, T, Boll, V, Hofmann, K, Baumann, U. | | Deposit date: | 2023-02-21 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Functional and structural diversity in deubiquitinases of the Chlamydia-like bacterium Simkania negevensis.
Nat Commun, 14, 2023
|
|
9F5T
 
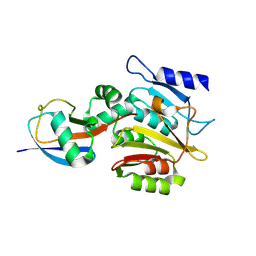 | | Ubiquitin C-terminal clippase BpJOS | | Descriptor: | BpJOS, Ubiquitin | | Authors: | Uthoff, M, Hermanns, T, Birkmann, S, Hofmann, K, Baumann, U. | | Deposit date: | 2024-04-30 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A family of bacterial Josephin-like deubiquitinases with an irreversible cleavage mode.
Mol.Cell, 85, 2025
|
|
6FSM
 
 | | Crystal structure of TCE-treated Thermolysin | | Descriptor: | CALCIUM ION, GLYCEROL, LYSINE, ... | | Authors: | Pichlo, C, Baumann, U. | | Deposit date: | 2018-02-19 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Improved protein-crystal identification by using 2,2,2-trichloroethanol as a fluorescence enhancer.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6GCO
 
 | | Truncated FtsH from A. aeolicus in P312 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent zinc metalloprotease FtsH, ZINC ION | | Authors: | Uthoff, M, Baumann, U. | | Deposit date: | 2018-04-18 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.323 Å) | | Cite: | Conformational flexibility of pore loop-1 gives insights into substrate translocation by the AAA+protease FtsH.
J. Struct. Biol., 204, 2018
|
|
1QDE
 
 | |
7NUU
 
 | | Crystal structure of human AMDHD2 in complex with Zn | | Descriptor: | GLYCEROL, N-acetylglucosamine-6-phosphate deacetylase, ZINC ION | | Authors: | Ruegenberg, S, Kroef, V, Baumann, U, Denzel, M.S. | | Deposit date: | 2021-03-14 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.836 Å) | | Cite: | GFPT2/GFAT2 and AMDHD2 act in tandem to control the hexosamine pathway.
Elife, 11, 2022
|
|
7NUT
 
 | | Crystal structure of human AMDHD2 in complex with Zn and GlcN6P | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, N-acetylglucosamine-6-phosphate deacetylase, ZINC ION | | Authors: | Ruegenberg, S, Kroef, V, Baumann, U, Denzel, M.S. | | Deposit date: | 2021-03-14 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | GFPT2/GFAT2 and AMDHD2 act in tandem to control the hexosamine pathway.
Elife, 11, 2022
|
|
