1YQK
 
 | | Human 8-oxoguanine glycosylase crosslinked with guanine containing DNA | | Descriptor: | 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*G)-3', 5'-D(P*CP*AP*GP*GP*TP*CP*TP*AP*C)-3', CALCIUM ION, ... | | Authors: | Banerjee, A, Yang, W, Karplus, M, Verdine, G.L. | | Deposit date: | 2005-02-01 | | Release date: | 2005-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA.
Nature, 434, 2005
|
|
1YQR
 
 | | Catalytically inactive human 8-oxoguanine glycosylase crosslinked to oxoG containing DNA | | Descriptor: | 5'-D(P*CP*GP*TP*CP*CP*AP*(8OG)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(P*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*G)-3', CALCIUM ION, ... | | Authors: | Banerjee, A, Yang, W, Karplus, M, Verdine, G.L. | | Deposit date: | 2005-02-02 | | Release date: | 2005-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA.
Nature, 434, 2005
|
|
1YQM
 
 | | Catalytically inactive human 8-oxoguanine glycosylase crosslinked to 7-deazaguanine containing DNA | | Descriptor: | 5'-D(P*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*C)-3', 5'-D(P*GP*TP*CP*CP*AP*(7GU)P*GP*TP*CP*TP*AP*C)-3', CALCIUM ION, ... | | Authors: | Banerjee, A, Yang, W, Karplus, M, Verdine, G.L. | | Deposit date: | 2005-02-02 | | Release date: | 2005-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA.
Nature, 434, 2005
|
|
1YQL
 
 | | Catalytically inactive hOGG1 crosslinked with 7-deaza-8-azaguanine containing DNA | | Descriptor: | 5'-D(P*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*C)-3', 5'-D(P*GP*TP*CP*CP*AP*(PPW)P*GP*TP*CP*TP*AP*C)-3', CALCIUM ION, ... | | Authors: | Banerjee, A, Yang, W, Karplus, M, Verdine, G.L. | | Deposit date: | 2005-02-02 | | Release date: | 2005-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA.
Nature, 434, 2005
|
|
6N0T
 
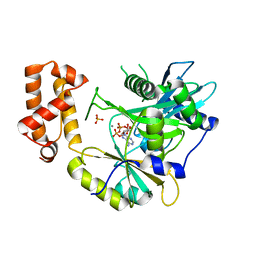 | | tRNA ligase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Banerjee, A, Goldgur, Y, Shuman, S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.511 Å) | | Cite: | Structure and two-metal mechanism of fungal tRNA ligase.
Nucleic Acids Res., 47, 2019
|
|
6N0V
 
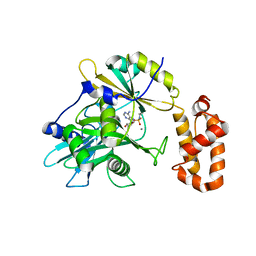 | | tRNA ligase | | Descriptor: | MANGANESE (II) ION, tRNA ligase | | Authors: | Banerjee, A, Goldgur, Y, Shuman, S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-01-09 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structure and two-metal mechanism of fungal tRNA ligase.
Nucleic Acids Res., 47, 2019
|
|
2I5W
 
 | |
4JTD
 
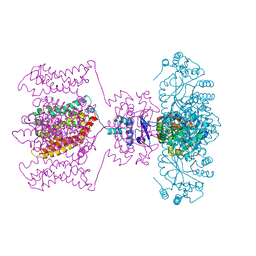 | | Crystal structure of Kv1.2-2.1 paddle chimera channel in complex with Lys27Met mutant of Charybdotoxin | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, ... | | Authors: | Banerjee, A, Lee, A, Campbell, E, MacKinnon, R. | | Deposit date: | 2013-03-23 | | Release date: | 2013-06-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structure of a pore-blocking toxin in complex with a eukaryotic voltage-dependent K(+) channel.
Elife, 2, 2013
|
|
5A8B
 
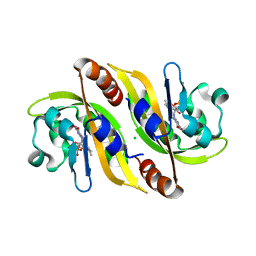 | | Structure of a parallel dimer of the aureochrome 1a LOV domain from Phaeodactylum tricornutum | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Banerjee, A, Herman, E, Kottke, T, Essen, L.O. | | Deposit date: | 2015-07-14 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.791 Å) | | Cite: | Structure of a Native-Like Aureochrome 1A Lov Domain Dimer from Phaeodactylum Tricornutum.
Structure, 24, 2016
|
|
4JTC
 
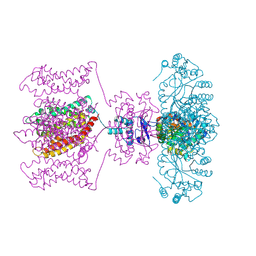 | | Crystal structure of Kv1.2-2.1 paddle chimera channel in complex with Charybdotoxin in Cs+ | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Banerjee, A, Lee, A, Campbell, E, MacKinnon, R. | | Deposit date: | 2013-03-23 | | Release date: | 2013-06-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure of a pore-blocking toxin in complex with a eukaryotic voltage-dependent K(+) channel.
Elife, 2, 2013
|
|
2F5O
 
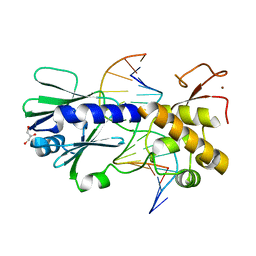 | | MutM crosslinked to undamaged DNA sampling G:C base pair IC3 | | Descriptor: | 5'-D(*AP*GP*GP*TP*AP*GP*AP*TP*CP*CP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*C*GP*TP*CP*CP*GP*GP*AP*TP*CP*TP*AP*CP*C)-3', GLYCEROL, ... | | Authors: | Banerjee, A, Santos, W.L, Verdine, G.L. | | Deposit date: | 2005-11-26 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of a DNA glycosylase searching for lesions.
Science, 311, 2006
|
|
2F5S
 
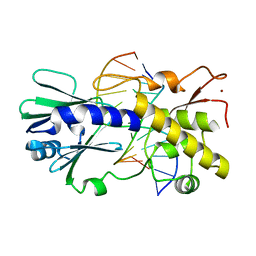 | | Catalytically inactive (E3Q) MutM crosslinked to oxoG:C containing DNA CC1 | | Descriptor: | 5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*TP*CP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*C*GP*TP*CP*CP*(8OG)P*AP*GP*TP*CP*TP*AP*CP*C)-3', ZINC ION, ... | | Authors: | Banerjee, A, Santos, W.L, Verdine, G.L. | | Deposit date: | 2005-11-26 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of a DNA glycosylase searching for lesions.
Science, 311, 2006
|
|
2F5Q
 
 | | Catalytically inactive (E3Q) MutM crosslinked to oxoG:C containing DNA CC2 | | Descriptor: | 5'-D(*AP*GP*G*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*CP*G*TP*CP*CP*AP*(8OG)P*GP*TP*CP*TP*AP*CP*C)-3', ZINC ION, ... | | Authors: | Banerjee, A, Santos, W.L, Verdine, G.L. | | Deposit date: | 2005-11-26 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of a DNA glycosylase searching for lesions.
Science, 311, 2006
|
|
2F5P
 
 | | MutM crosslinked to undamaged DNA sampling A:T base pair IC2 | | Descriptor: | 5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*TP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*CP*G*TP*CP*CP*AP*AP*GP*TP*CP*TP*AP*CP*C)-3', ZINC ION, ... | | Authors: | Banerjee, A, Santos, W.L, Verdine, G.L. | | Deposit date: | 2005-11-26 | | Release date: | 2006-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a DNA glycosylase searching for lesions.
Science, 311, 2006
|
|
2F5N
 
 | | MutM crosslinked to undamaged DNA sampling A:T base pair IC1 | | Descriptor: | 5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*C*GP*TP*CP*CP*AP*GP*GP*TP*CP*TP*AP*CP*C)-3', GLYCEROL, ... | | Authors: | Banerjee, A, Santos, W.L, Verdine, G.L. | | Deposit date: | 2005-11-26 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a DNA glycosylase searching for lesions.
Science, 311, 2006
|
|
8YP6
 
 | |
8P8M
 
 | | Yeast 60S ribosomal subunit, RPL39 deletion | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Rabl, J, Banerjee, A, Boehringer, D, Zavolan, M. | | Deposit date: | 2023-06-01 | | Release date: | 2024-06-12 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Ribosomal protein RPL39L is an efficiency factor in the cotranslational folding of a subset of proteins with alpha helical domains.
Nucleic Acids Res., 52, 2024
|
|
8PFR
 
 | | Mouse RPL39L integrated into the yeast 60S ribosomal subunit | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Rabl, J, Banerjee, A, Boehringer, D, Zavolan, M. | | Deposit date: | 2023-06-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Ribosomal protein RPL39L is an efficiency factor in the cotranslational folding of a subset of proteins with alpha helical domains.
Nucleic Acids Res., 52, 2024
|
|
8P8U
 
 | | Yeast 60S ribosomal subunit | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Rabl, J, Banerjee, A, Boehringer, D, Zavolan, M. | | Deposit date: | 2023-06-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | Ribosomal protein RPL39L is an efficiency factor in the cotranslational folding of a subset of proteins with alpha helical domains.
Nucleic Acids Res., 52, 2024
|
|
8P8N
 
 | | Mouse RPL39 integrated into the yeast 60S ribosomal subunit | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Rabl, J, Banerjee, A, Boehringer, D, Zavolan, M. | | Deposit date: | 2023-06-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Ribosomal protein RPL39L is an efficiency factor in the cotranslational folding of a subset of proteins with alpha helical domains.
Nucleic Acids Res., 52, 2024
|
|
6U05
 
 | | Crystal Structure of Fungal RNA Kinase | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Shuman, S, Goldgur, Y, Banerjee, A. | | Deposit date: | 2019-08-13 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Atomic structures of the RNA end-healing 5'-OH kinase and 2',3'-cyclic phosphodiesterase domains of fungal tRNA ligase: conformational switches in the kinase upon binding of the GTP phosphate donor.
Nucleic Acids Res., 47, 2019
|
|
6TZX
 
 | | Crystal Structure of Fungal RNA Kinase | | Descriptor: | INOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, tRNA ligase | | Authors: | Shuman, S, Goldgur, Y, Banerjee, A. | | Deposit date: | 2019-08-13 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.529 Å) | | Cite: | Atomic structures of the RNA end-healing 5'-OH kinase and 2',3'-cyclic phosphodiesterase domains of fungal tRNA ligase: conformational switches in the kinase upon binding of the GTP phosphate donor.
Nucleic Acids Res., 47, 2019
|
|
6U03
 
 | | Crystal Structure of Fungal RNA Kinase | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, tRNA ligase | | Authors: | Shuman, S, Goldgur, Y, Banerjee, A. | | Deposit date: | 2019-08-13 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Atomic structures of the RNA end-healing 5'-OH kinase and 2',3'-cyclic phosphodiesterase domains of fungal tRNA ligase: conformational switches in the kinase upon binding of the GTP phosphate donor.
Nucleic Acids Res., 47, 2019
|
|
6U00
 
 | | Crystal Structure of Fungal RNA Kinase | | Descriptor: | PHOSPHATE ION, tRNA ligase | | Authors: | Shuman, S, Goldgur, Y, Banerjee, A. | | Deposit date: | 2019-08-13 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Atomic structures of the RNA end-healing 5'-OH kinase and 2',3'-cyclic phosphodiesterase domains of fungal tRNA ligase: conformational switches in the kinase upon binding of the GTP phosphate donor.
Nucleic Acids Res., 47, 2019
|
|
6TZO
 
 | | Crystal Structure of Fungal RNA Kinase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, tRNA ligase | | Authors: | Shuman, S, Goldgur, Y, Banerjee, A. | | Deposit date: | 2019-08-12 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Atomic structures of the RNA end-healing 5'-OH kinase and 2',3'-cyclic phosphodiesterase domains of fungal tRNA ligase: conformational switches in the kinase upon binding of the GTP phosphate donor.
Nucleic Acids Res., 47, 2019
|
|
