3ZBO
 
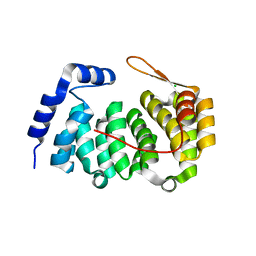 | | A new family of proteins related to the HEAT-like repeat DNA glycosylases with affinity for branched DNA structures | | Descriptor: | ALKF, CHLORIDE ION | | Authors: | Backe, P.H, Simm, R, Laerdahl, J.K, Dalhus, B, Fagerlund, A, Okstad, O.A, Rognes, T, Alseth, I, Kolsto, A.-B, Bjoras, M. | | Deposit date: | 2012-11-12 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A New Family of Proteins Related to the Heat-Like Repeat DNA Glycosylases with Affinity for Branched DNA Structures.
J.Struct.Biol., 183, 2013
|
|
6SNP
 
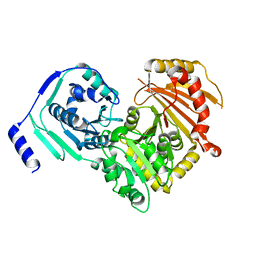 | | Crystal structures of human PGM1 isoform 2 | | Descriptor: | MAGNESIUM ION, Phosphoglucomutase-1 | | Authors: | Backe, P.H, Laerdahl, J.K, Kittelsen, L.S, Dalhus, B, Morkrid, L, Bjoras, M. | | Deposit date: | 2019-08-27 | | Release date: | 2020-04-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for substrate and product recognition in human phosphoglucomutase-1 (PGM1) isoform 2, a member of the alpha-D-phosphohexomutase superfamily.
Sci Rep, 10, 2020
|
|
6SNO
 
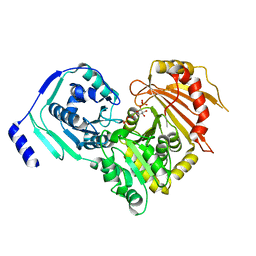 | | Crystal structures of human PGM1 isoform 2 | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, Phosphoglucomutase-1, ZINC ION | | Authors: | Backe, P.H, Laerdahl, J.K, Kittelsen, L.S, Dalhus, B, Morkrid, L, Bjoras, M. | | Deposit date: | 2019-08-27 | | Release date: | 2020-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for substrate and product recognition in human phosphoglucomutase-1 (PGM1) isoform 2, a member of the alpha-D-phosphohexomutase superfamily.
Sci Rep, 10, 2020
|
|
6SNQ
 
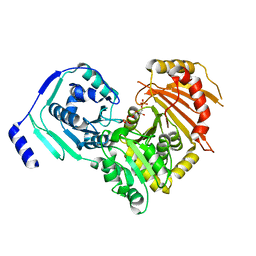 | | Crystal structures of human PGM1 isoform 2 | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, Phosphoglucomutase-1, ZINC ION | | Authors: | Backe, P.H, Laerdahl, J.K, Kittelsen, L.S, Dalhus, B, Morkrid, L, Bjoras, M. | | Deposit date: | 2019-08-27 | | Release date: | 2020-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for substrate and product recognition in human phosphoglucomutase-1 (PGM1) isoform 2, a member of the alpha-D-phosphohexomutase superfamily.
Sci Rep, 10, 2020
|
|
1ZZI
 
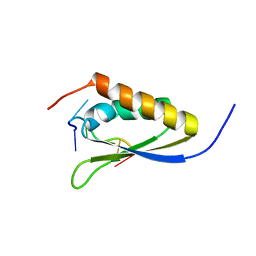 | | Crystal Structure Analysis of the third KH domain of hnRNP K in complex with ssDNA | | Descriptor: | 5'-D(*CP*TP*CP*CP*CP*C)-3', Heterogeneous nuclear ribonucleoprotein K | | Authors: | Backe, P.H, Messias, A.C, Ravelli, R.B, Sattler, M, Cusack, S. | | Deposit date: | 2005-06-14 | | Release date: | 2005-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Crystallographic and NMR Studies of the Third KH Domain of hnRNP K in Complex with Single-Stranded Nucleic Acids
STRUCTURE, 13, 2005
|
|
1ZZJ
 
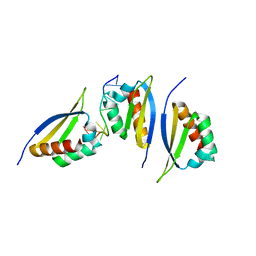 | | Structure of the third KH domain of hnRNP K in complex with 15-mer ssDNA | | Descriptor: | 5'-D(*TP*TP*CP*CP*CP*CP*TP*CP*CP*CP*CP*AP*TP*TP*T)-3', Heterogeneous nuclear ribonucleoprotein K | | Authors: | Backe, P.H, Messias, A.C, Ravelli, R.B, Sattler, M, Cusack, S. | | Deposit date: | 2005-06-14 | | Release date: | 2005-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-Ray Crystallographic and NMR Studies of the Third KH Domain of hnRNP K in Complex with Single-Stranded Nucleic Acids
STRUCTURE, 13, 2005
|
|
1ZZK
 
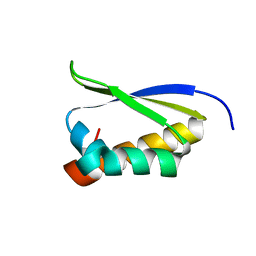 | | Crystal Structure of the third KH domain of hnRNP K at 0.95A resolution | | Descriptor: | Heterogeneous nuclear ribonucleoprotein K | | Authors: | Backe, P.H, Messias, A.C, Ravelli, R.B, Sattler, M, Cusack, S. | | Deposit date: | 2005-06-14 | | Release date: | 2005-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | X-Ray Crystallographic and NMR Studies of the Third KH Domain of hnRNP K in Complex with Single-Stranded Nucleic Acids
STRUCTURE, 13, 2005
|
|
4AXN
 
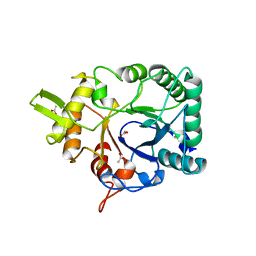 | | Hallmarks of processive and non-processive glycoside hydrolases revealed from computational and crystallographic studies of the Serratia marcescens chitinases | | Descriptor: | ACETATE ION, CALCIUM ION, CHITINASE C1 | | Authors: | Payne, C.M, Baban, J, Synstad, B, Backe, P.H, Arvai, A.S, Dalhus, B, Bjoras, M, Eijsink, V.G.H, Sorlie, M, Beckham, G.T, Vaaje-Kolstad, G. | | Deposit date: | 2012-06-13 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Hallmarks of Processivity in Glycoside Hydrolases from Crystallographic and Computational Studies of the Serratia Marcescens Chitinases.
J.Biol.Chem., 287, 2012
|
|
4AE6
 
 | | Structure and Function of the Human Sperm-Specific Isoform of Protein Kinase A (PKA) Catalytic Subunit Calpha 2 | | Descriptor: | ACETATE ION, CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA 2 | | Authors: | Hereng, T.H, Backe, P.H, Kahmann, J, Scheich, C, Bjoras, M, Skalhegg, B.S, Rosendal, K.R. | | Deposit date: | 2012-01-09 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Function of the Human Sperm-Specific Isoform of Protein Kinase a (Pka) Catalytic Subunit Calpha2
J.Struct.Biol., 178, 2012
|
|
2XHI
 
 | | Separation-of-function mutants unravel the dual reaction mode of human 8-oxoguanine DNA glycosylase | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*AP*(8OG)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Dalhus, B, Forsbring, M, Helle, I.H, Backe, P.H, Forstrom, R.J, Alseth, I, Bjoras, M. | | Deposit date: | 2010-06-16 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Separation-of-Function Mutants Unravel the Dual- Reaction Mode of Human 8-Oxoguanine DNA Glycosylase.
Structure, 19, 2011
|
|
2W36
 
 | | Structures of endonuclease V with DNA reveal initiation of deaminated adenine repair | | Descriptor: | 5'-D(*CP*GP*AP*TP*CP*TP*GP*TP*AP*GP*CP)-3', 5'-D(*GP*CP*BRUP*AP*CP*IP*GP*AP*BRUP*CP*GP)-3', ENDONUCLEASE V | | Authors: | Dalhus, B, Arvai, A.S, Rosnes, I, Olsen, O.E, Backe, P.H, Alseth, I, Gao, H, Cao, W, Tainer, J.A, Bjoras, M. | | Deposit date: | 2008-11-06 | | Release date: | 2009-01-20 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Endonuclease V with DNA Reveal Initiation of Deaminated Adenine Repair.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2W35
 
 | | Structures of endonuclease V with DNA reveal initiation of deaminated adenine repair | | Descriptor: | 5'-D(*AP*GP*CP*CP*GP*TP)-3', 5'-D(*AP*TP*GP*CP*GP*AP*CP*IP*GP)-3', Endonuclease V, ... | | Authors: | Dalhus, B, Arvai, A.S, Rosnes, I, Olsen, O.E, Backe, P.H, Alseth, I, Gao, H, Cao, W, Tainer, J.A, Bjoras, M. | | Deposit date: | 2008-11-06 | | Release date: | 2009-01-20 | | Last modified: | 2023-07-05 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of Endonuclease V with DNA Reveal Initiation of Deaminated Adenine Repair.
Nat.Struct.Mol.Biol., 16, 2009
|
|
4AE9
 
 | | Structure and function of the Human Sperm-Specific Isoform of Protein Kinase A (PKA) Catalytic Subunit C alpha 2 | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA | | Authors: | Hereng, T.H, Backe, P.H, Kahmann, J, Scheich, C, Bjoras, M, Skalhegg, B.S, Rosendal, K.R. | | Deposit date: | 2012-01-09 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Function of the Human Sperm-Specific Isoform of Protein Kinase a (Pka) Catalytic Subunit Calpha2
J.Struct.Biol., 178, 2012
|
|
