4I4J
 
 | | The structure of SgcE10, the ACP-polyene thioesterase involved in C-1027 biosynthesis | | 分子名称: | 1,2-ETHANEDIOL, ACP-polyene thioesterase, D(-)-TARTARIC ACID, ... | | 著者 | Kim, Y, Bigelow, L, Bearden, J, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2012-11-27 | | 公開日 | 2012-12-12 | | 最終更新日 | 2022-05-04 | | 実験手法 | X-RAY DIFFRACTION (2.784 Å) | | 主引用文献 | Crystal Structure of Thioesterase SgcE10 Supporting Common Polyene Intermediates in 9- and 10-Membered Enediyne Core Biosynthesis.
Acs Omega, 2, 2017
|
|
5I4Q
 
 | | Contact-dependent inhibition system from Escherichia coli NC101 - ternary CdiA/CdiI/EF-Tu complex (domains 2 and 3) | | 分子名称: | CHLORIDE ION, Contact-dependent inhibitor A, Contact-dependent inhibitor I, ... | | 著者 | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | 登録日 | 2016-02-12 | | 公開日 | 2017-06-28 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structure of a novel antibacterial toxin that exploits elongation factor Tu to cleave specific transfer RNAs.
Nucleic Acids Res., 45, 2017
|
|
5I4R
 
 | | Contact-dependent inhibition system from Escherichia coli NC101 - ternary CdiA/CdiI/EF-Tu complex (trypsin-modified) | | 分子名称: | Contact-dependent inhibitor A, Contact-dependent inhibitor I, Elongation factor Tu, ... | | 著者 | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | 登録日 | 2016-02-12 | | 公開日 | 2017-06-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structure of a novel antibacterial toxin that exploits elongation factor Tu to cleave specific transfer RNAs.
Nucleic Acids Res., 45, 2017
|
|
4QYR
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsE KS3 | | 分子名称: | ACETIC ACID, AT-less polyketide synthase, CHLORIDE ION, ... | | 著者 | Kim, Y, Li, H, Endres, M, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2014-07-25 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.902 Å) | | 主引用文献 | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4F66
 
 | | The crystal structure of 6-phospho-beta-glucosidase from Streptococcus mutans UA159 in complex with beta-D-glucose-6-phosphate. | | 分子名称: | 1,2-ETHANEDIOL, 6-O-phosphono-beta-D-glucopyranose, FORMIC ACID, ... | | 著者 | Tan, K, Michalska, K, Hatzos-Skintges, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-05-14 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.479 Å) | | 主引用文献 | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
4F79
 
 | | The crystal structure of 6-phospho-beta-glucosidase mutant (E375Q) in complex with Salicin 6-phosphate | | 分子名称: | 2-(hydroxymethyl)phenyl 6-O-phosphono-beta-D-glucopyranoside, GLYCEROL, Putative phospho-beta-glucosidase | | 著者 | Tan, K, Michalska, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-05-15 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
4GPN
 
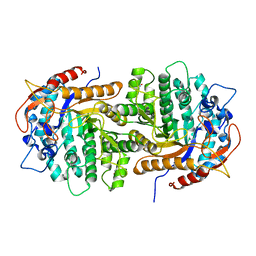 | | The crystal structure of 6-P-beta-D-Glucosidase (E375Q mutant) from Streptococcus mutans UA150 in complex with Gentiobiose 6-phosphate. | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-O-phosphono-beta-D-glucopyranose-(1-6)-beta-D-glucopyranose, 6-phospho-beta-D-Glucosidase, ... | | 著者 | Tan, K, Michalska, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-08-21 | | 公開日 | 2012-10-03 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.291 Å) | | 主引用文献 | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
4GZE
 
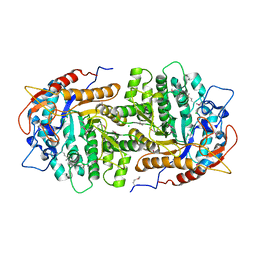 | | Crystal structure of 6-phospho-beta-glucosidase from Lactobacillus plantarum (apo form) | | 分子名称: | 6-phospho-beta-glucosidase, CHLORIDE ION, GLYCEROL | | 著者 | Michalska, K, Hatzos-Skintges, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-09-06 | | 公開日 | 2012-09-26 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4O2H
 
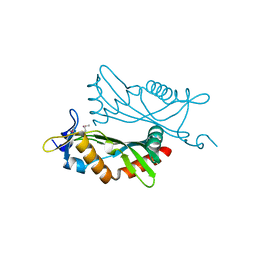 | | Crystal structure of BCAM1869 protein (RsaM homolog) from Burkholderia cenocepacia | | 分子名称: | protein BCAM1869 | | 著者 | Michalska, K, Chhor, G, Clancy, S, Winans, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-12-17 | | 公開日 | 2014-01-22 | | 最終更新日 | 2014-10-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | RsaM: a transcriptional regulator of Burkholderia spp. with novel fold.
Febs J., 281, 2014
|
|
6WRH
 
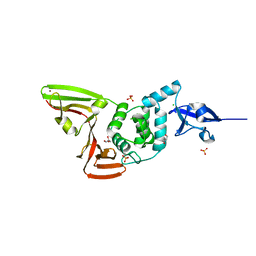 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant | | 分子名称: | CHLORIDE ION, GLYCEROL, Non-structural protein 3, ... | | 著者 | Osipiuk, J, Tesar, C, Jedrzejczak, R, Endres, M, Welk, L, Babnigg, G, Kim, Y, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-29 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
4DWJ
 
 | | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in complex with Thymidine Monophosphate | | 分子名称: | THYMIDINE-5'-PHOSPHATE, Thymidylate kinase | | 著者 | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | 登録日 | 2012-02-24 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.74 Å) | | 主引用文献 | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in complex with Thymidine Monophosphate
To be Published
|
|
4EAQ
 
 | | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in complex with 3'-Azido-3'-Deoxythymidine-5'-Monophosphate | | 分子名称: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-MONOPHOSPHATE, Thymidylate kinase | | 著者 | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | 登録日 | 2012-03-22 | | 公開日 | 2012-04-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in complex with 3'-Azido-3'-Deoxythymidine-5'-Monophosphate
To be Published
|
|
3OON
 
 | | The structure of an outer membrance protein from Borrelia burgdorferi B31 | | 分子名称: | Outer membrane protein (Tpn50), SULFATE ION | | 著者 | Fan, Y, Bigelow, L, Feldman, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-08-31 | | 公開日 | 2010-09-22 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Insights into PG-binding, conformational change, and dimerization of the OmpA C-terminal domains from Salmonella enterica serovar Typhimurium and Borrelia burgdorferi.
Protein Sci., 26, 2017
|
|
3QOM
 
 | | Crystal structure of 6-phospho-beta-glucosidase from Lactobacillus plantarum | | 分子名称: | 6-phospho-beta-glucosidase, ACETATE ION, PHOSPHATE ION, ... | | 著者 | Michalska, K, Hatzos-Skintges, C, Bearden, J, Kohler, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-02-10 | | 公開日 | 2011-03-09 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.498 Å) | | 主引用文献 | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4ZDN
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsF KS4 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AT-less polyketide synthase, CHLORIDE ION | | 著者 | Chang, C, Li, H, Endres, M, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-04-17 | | 公開日 | 2015-05-13 | | 最終更新日 | 2023-03-22 | | 実験手法 | X-RAY DIFFRACTION (2.509 Å) | | 主引用文献 | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZWV
 
 | | Crystal Structure of Aminotransferase AtmS13 from Actinomadura melliaura | | 分子名称: | GLYCEROL, Putative aminotransferase | | 著者 | Kim, Y, Bigelow, L, Endres, M, Wang, F, Phillips Jr, G.N, Joachimiak, A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-05-19 | | 公開日 | 2015-06-03 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (1.503 Å) | | 主引用文献 | Structural characterization of AtmS13, a putative sugar aminotransferase involved in indolocarbazole AT2433 aminopentose biosynthesis.
Proteins, 83, 2015
|
|
5DN1
 
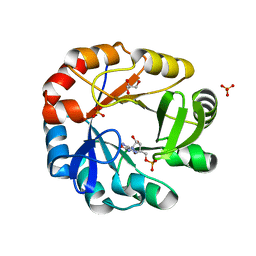 | | Crystal structure of Phosphoribosyl isomerase A from Streptomyces coelicolor | | 分子名称: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, GLYCEROL, Phosphoribosyl isomerase A, ... | | 著者 | Chang, C, Verduzco-Castro, E.A, Endres, M, Barona-Gomez, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-09-09 | | 公開日 | 2015-09-30 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.953 Å) | | 主引用文献 | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
3UO3
 
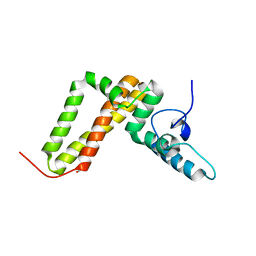 | | Jac1 co-chaperone from Saccharomyces cerevisiae, 5-182 clone | | 分子名称: | ACETATE ION, J-type co-chaperone JAC1, mitochondrial | | 著者 | Osipiuk, J, Bigelow, L, Mulligan, R, Feldmann, B, Babnigg, G, Marszalek, J, Craig, E.A, Dutkiewicz, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-11-16 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Interaction of j-protein co-chaperone jac1 with fe-s scaffold isu is indispensable in vivo and conserved in evolution.
J.Mol.Biol., 417, 2012
|
|
4JGP
 
 | | The crystal structure of sporulation kinase D sensor domain from Bacillus subtilis subsp in complex with pyruvate at 2.0A resolution | | 分子名称: | PYRUVIC ACID, Sporulation kinase D | | 著者 | Wu, R, Schiffer, M, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-03-01 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Insight into the sporulation phosphorelay: Crystal structure of the sensor domain of Bacillus subtilis histidine kinase, KinD.
Protein Sci., 22, 2013
|
|
4JGQ
 
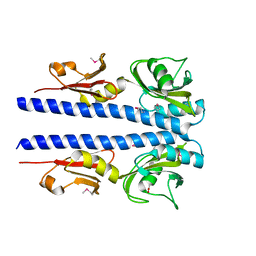 | | The crystal structure of sporulation kinase D mutant sensor domain, r131a, from Bacillus subtilis subsp in co-crystallization with pyruvate | | 分子名称: | ACETIC ACID, Sporulation kinase D | | 著者 | Wu, R, Schiffer, M, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-03-01 | | 公開日 | 2013-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | Insight into the sporulation phosphorelay: Crystal structure of the sensor domain of Bacillus subtilis histidine kinase, KinD.
Protein Sci., 22, 2013
|
|
4JGO
 
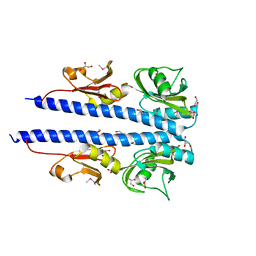 | | The crystal structure of sporulation kinase d sensor domain from Bacillus subtilis subsp. | | 分子名称: | GLYCEROL, PYRUVIC ACID, Sporulation kinase D | | 著者 | Wu, R, Schiffer, M, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-03-01 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Insight into the sporulation phosphorelay: Crystal structure of the sensor domain of Bacillus subtilis histidine kinase, KinD.
Protein Sci., 22, 2013
|
|
4JGR
 
 | | The crystal structure of sporulation kinase D mutant sensor domain, R131A, from Bacillus subtilis subsp at 2.4A resolution | | 分子名称: | ACETIC ACID, GLYCEROL, Sporulation kinase D | | 著者 | Wu, R, Schiffer, M, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-03-01 | | 公開日 | 2013-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Insight into the sporulation phosphorelay: Crystal structure of the sensor domain of Bacillus subtilis histidine kinase, KinD.
Protein Sci., 22, 2013
|
|
4K08
 
 | | Periplasmic sensor domain of chemotaxis protein, Adeh_3718 | | 分子名称: | ACETATE ION, Methyl-accepting chemotaxis sensory transducer, ZINC ION | | 著者 | Pokkuluri, P.R, Mack, J.C, Bearden, J, Rakowski, E, Schiffer, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-04-03 | | 公開日 | 2013-07-17 | | 最終更新日 | 2015-04-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Analysis of periplasmic sensor domains from Anaeromyxobacter dehalogenans 2CP-C: structure of one sensor domain from a histidine kinase and another from a chemotaxis protein.
Microbiologyopen, 2, 2013
|
|
4K0D
 
 | | Periplasmic sensor domain of sensor histidine kinase, Adeh_2942 | | 分子名称: | ACETATE ION, CHLORIDE ION, Periplasmic sensor hybrid histidine kinase, ... | | 著者 | Pokkuluri, P.R, Mack, J.C, Bearden, J, Rakowski, E, Schiffer, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-04-03 | | 公開日 | 2013-06-12 | | 最終更新日 | 2015-04-15 | | 実験手法 | X-RAY DIFFRACTION (1.997 Å) | | 主引用文献 | Analysis of periplasmic sensor domains from Anaeromyxobacter dehalogenans 2CP-C: structure of one sensor domain from a histidine kinase and another from a chemotaxis protein.
Microbiologyopen, 2, 2013
|
|
4OO2
 
 | | Streptomyces globisporus C-1027 FAD dependent (S)-3-chloro-β-tyrosine-S-SgcC2 C-5 hydroxylase SgcC apo form | | 分子名称: | CALCIUM ION, Chlorophenol-4-monooxygenase, GLYCEROL | | 著者 | Cao, H, Xu, W, Bingman, C.A, Lohman, J.R, Yennamalli, R, Shen, B, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2014-01-29 | | 公開日 | 2014-02-12 | | 最終更新日 | 2023-03-22 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
