4EBW
 
 | | Structure of Focal Adhesion Kinase catalytic domain in complex with novel allosteric inhibitor | | Descriptor: | 1-ethyl-8-(4-ethylphenyl)-5-methyl-1,5-dihydropyrazolo[4,3-c][2,1]benzothiazine 4,4-dioxide, Focal adhesion kinase 1 | | Authors: | Iwatani, M, Iwata, H, Okabe, A, Skene, R.J, Tomita, N, Hayashi, Y, Aramaki, Y, Hosfield, D.J, Hori, A, Baba, A, Miki, H. | | Deposit date: | 2012-03-25 | | Release date: | 2012-07-25 | | Last modified: | 2013-03-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery and characterization of novel allosteric FAK inhibitors.
Eur.J.Med.Chem., 61, 2013
|
|
1LR1
 
 | | Solution Structure of the Oligomerization Domain of the Bacterial Chromatin-Structuring Protein H-NS | | Descriptor: | dna-binding protein h-ns | | Authors: | Esposito, D, Petrovic, A, Harris, R, Ono, S, Eccleston, J, Mbabaali, A, Haq, I, Higgins, C.F, Hinton, J.C.D, Driscoll, P.C, Ladbury, J.E. | | Deposit date: | 2002-05-14 | | Release date: | 2003-01-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | H-NS Oligomerization Domain Structure Reveals the Mechanism for High Order
Self-association of the Intact Protein
J.Mol.Biol., 324, 2002
|
|
6O3Z
 
 | | Crystal structure of RORgt with 3-cyano-N-(3-{[(3S)-4-(cyclopentanecarbonyl)-3-methylpiperazin-1-yl]methyl}-5-fluoro-2-methylphenyl)benzamide (compound 1) | | Descriptor: | 1,2-ETHANEDIOL, 3-cyano-N-(3-{[(3S)-4-(cyclopentanecarbonyl)-3-methylpiperazin-1-yl]methyl}-5-fluoro-2-methylphenyl)benzamide, RAR-related orphan receptor C isoform a variant | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of [1,2,4]Triazolo[1,5-a]pyridine Derivatives as Potent and Orally Bioavailable ROR gamma t Inverse Agonists.
Acs Med.Chem.Lett., 11, 2020
|
|
4EBV
 
 | | Structure of Focal Adhesion Kinase catalytic domain in complex with novel allosteric inhibitor | | Descriptor: | 8-(4-ethylphenyl)-5-methyl-2,5-dihydropyrazolo[4,3-c][2,1]benzothiazine 4,4-dioxide, Focal adhesion kinase 1, ISOPROPYL ALCOHOL | | Authors: | Skene, R.J, Hosfield, D.J. | | Deposit date: | 2012-03-25 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery and characterization of novel allosteric FAK inhibitors.
Eur.J.Med.Chem., 61, 2013
|
|
4I4E
 
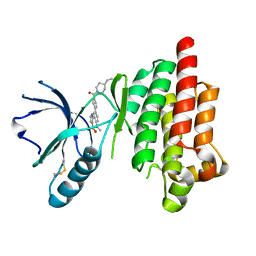 | | Structure of Focal Adhesion Kinase catalytic domain in complex with hinge binding pyrazolobenzothiazine compound. | | Descriptor: | Focal adhesion kinase 1, [4-(2-hydroxyethyl)piperidin-1-yl][4-(5-methyl-4,4-dioxido-1,5-dihydropyrazolo[4,3-c][2,1]benzothiazin-8-yl)phenyl]methanone | | Authors: | Skene, R.J, Hosfield, D.J. | | Deposit date: | 2012-11-27 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-based discovery of cellular-active allosteric inhibitors of FAK.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4I4F
 
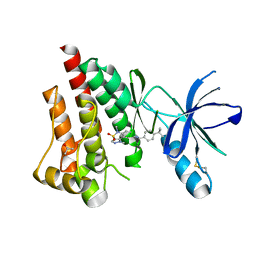 | | Structure of Focal Adhesion Kinase catalytic domain in complex with an allosteric binding pyrazolobenzothiazine compound. | | Descriptor: | Focal adhesion kinase 1, ISOPROPYL ALCOHOL, N-(4-tert-butylbenzyl)-1,5-dimethyl-1,5-dihydropyrazolo[4,3-c][2,1]benzothiazin-8-amine 4,4-dioxide | | Authors: | Skene, R.J, Hosfield, D.J. | | Deposit date: | 2012-11-27 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-based discovery of cellular-active allosteric inhibitors of FAK.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6OAM
 
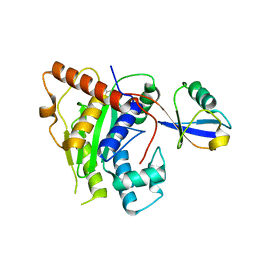 | | Crystal Structure of ChlaDUB2 DUB domain | | Descriptor: | Deubiquitinase and deneddylase Dub2, Ubiquitin | | Authors: | Hausman, J.M, Das, C. | | Deposit date: | 2019-03-17 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | The Two Deubiquitinating Enzymes fromChlamydia trachomatisHave Distinct Ubiquitin Recognition Properties.
Biochemistry, 59, 2020
|
|
2KUS
 
 | |
6MRN
 
 | | Crystal Structure of ChlaDUB2 DUB domain | | Descriptor: | Deubiquitinase and deneddylase Dub2 | | Authors: | Hausman, J.M, Das, C. | | Deposit date: | 2018-10-15 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The Two Deubiquitinating Enzymes fromChlamydia trachomatisHave Distinct Ubiquitin Recognition Properties.
Biochemistry, 59, 2020
|
|
7LM3
 
 | | Crystal Structure of Thr316Ala mutant of JAMM domain of S. pombe | | Descriptor: | AMSH-like protease sst2, PHOSPHATE ION, ZINC ION | | Authors: | Shrestha, R, Das, C. | | Deposit date: | 2021-02-05 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Thr316Ala mutant of a yeast JAMM deubiquitinase: implication of active-site loop dynamics in catalysis.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
