5TA4
 
 | | Discovery of a Potent Cyclophilin Inhibitor (Compound 8) based on Structural Simplification of Sanglifehrin A | | Descriptor: | 18-methoxy-2,11,17-trimethyl-14-(propan-2-yl)-3-oxa-9,12,15,28-tetraazatricyclo[21.3.1.1~5,9~]octacosa-1(27),21,23,25-tetraene-4,10,13,16-tetrone, Peptidyl-prolyl cis-trans isomerase A, SULFATE ION | | Authors: | Appleby, T.C, Steadman, V, Pettit, S, Schmitz, U, Mackman, R.L, Schultz, B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Potent Cyclophilin Inhibitors Based on the Structural Simplification of Sanglifehrin A.
J. Med. Chem., 60, 2017
|
|
6V2F
 
 | | Crystal structure of the HIV capsid hexamer bound to the small molecule long-acting inhibitor, GS-6207 | | Descriptor: | HIV-1 capsid, N-[(1S)-1-(3-{4-chloro-3-[(methylsulfonyl)amino]-1-(2,2,2-trifluoroethyl)-1H-indazol-7-yl}-6-[3-methyl-3-(methylsulfonyl)but-1-yn-1-yl]pyridin-2-yl)-2-(3,5-difluorophenyl)ethyl]-2-[(3bS,4aR)-5,5-difluoro-3-(trifluoromethyl)-3b,4,4a,5-tetrahydro-1H-cyclopropa[3,4]cyclopenta[1,2-c]pyrazol-1-yl]acetamide | | Authors: | Appleby, T.C, Link, J.O, Yant, S.R, Villasenor, A.G, Somoza, J.R, Hu, E.Y, Schroeder, S.D, Cihlar, T. | | Deposit date: | 2019-11-22 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Clinical targeting of HIV capsid protein with a long-acting small molecule.
Nature, 584, 2020
|
|
4WTD
 
 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS AND DELTA8 BETA HAIRPIN LOOP DELETION IN COMPLEX WITH ADP, MN2+ AND SYMMETRICAL PRIMER TEMPLATE 5'-AUAAAUUU | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Edwards, T.E, Fox III, D, Appleby, T.C. | | Deposit date: | 2014-10-29 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
4WTF
 
 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS AND DELTA8 BETA HAIRPIN LOOP DELETION IN COMPLEX WITH GS-639475, MN2+ AND SYMMETRICAL PRIMER TEMPLATE 5'-CAAAAUUU | | Descriptor: | 2'-C-methyluridine 5'-(trihydrogen diphosphate), CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Edwards, T.E, Appleby, T.C, Mcgrath, M.E. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
4WTJ
 
 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS IN COMPLEX WITH RNA TEMPLATE 5'-AUCC, RNA PRIMER 5'-PGG, MN2+, AND ADP | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Edwards, T.E, Appleby, T.C. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
4WTM
 
 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS IN COMPLEX WITH RNA TEMPLATE 5'-UAGG, RNA PRIMER 5'-PCC, MN2+, AND UDP | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, RNA PRIMER CC, ... | | Authors: | Edwards, T.E, Appleby, T.C, Fox III, D. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
4WT9
 
 | |
4WTA
 
 | |
4WTC
 
 | |
4WTE
 
 | |
4WTG
 
 | |
4WTI
 
 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS IN COMPLEX WITH RNA TEMPLATE 5'-ACGG, RNA PRIMER 5'-PCC, MN2+, AND GDP | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Edwards, T.E, Appleby, T.C. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
4WTK
 
 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS IN COMPLEX WITH RNA TEMPLATE 5'-AGCC, RNA PRIMER 5'-PGG, MN2+, AND CDP | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Edwards, T.E, Appleby, T.C. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
4WTL
 
 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS IN COMPLEX WITH RNA TEMPLATE 5'-UACC, RNA PRIMER 5'-PGG, MN2+, AND UDP | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Edwards, T.E, Appleby, T.C. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
7UO7
 
 | | SARS-CoV-2 replication-transcription complex bound to ATP, in a pre-catalytic state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
7UO4
 
 | | SARS-CoV-2 replication-transcription complex bound to Remdesivir triphosphate, in a pre-catalytic state | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
7UO9
 
 | | SARS-CoV-2 replication-transcription complex bound to UTP, in a pre-catalytic state | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
7UOE
 
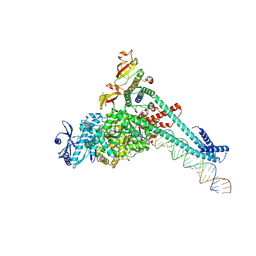 | | SARS-CoV-2 replication-transcription complex bound to CTP, in a pre-catalytic state | | Descriptor: | 3'-DEOXYURIDINE-5'-MONOPHOSPHATE, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
7UOB
 
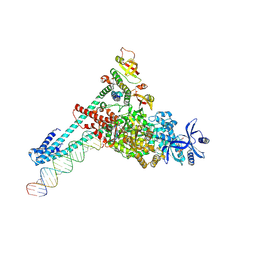 | | SARS-CoV-2 replication-transcription complex bound to GTP, in a pre-catalytic state | | Descriptor: | 3'-DEOXYURIDINE-5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
5UJ2
 
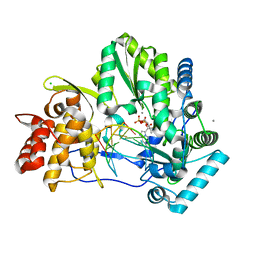 | | Crystal structure of HCV NS5B genotype 2A JFH-1 isolate with S15G E86Q E87Q C223H V321I mutations and Delta8 neta hairpoin loop deletion in complex with GS-639476 (diphsohate version of GS-9813), Mn2+ and symmetrical primer template 5'-AUAAAUUU | | Descriptor: | (1S)-1-(4-aminoimidazo[2,1-f][1,2,4]triazin-7-yl)-1,4-anhydro-2-deoxy-2-fluoro-5-O-[(S)-hydroxy(phosphonooxy)phosphoryl]-2-methyl-D-ribitol, CHLORIDE ION, Genome polyprotein, ... | | Authors: | Edwards, T.E, Fox III, D, Appleby, T.C, Murakami, E, Rey, A, McGrath, M.E. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of a 2'-fluoro-2'-C-methyl C-nucleotide HCV polymerase inhibitor and a phosphoramidate prodrug with favorable properties.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
4EO6
 
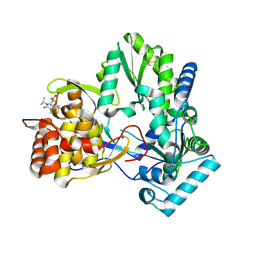 | |
4EO8
 
 | | HCV NS5B polymerase inhibitors: Tri-substituted acylhydrazines as tertiary amide bioisosteres | | Descriptor: | 5-(3,3-dimethylbut-1-yn-1-yl)-3-{2,2-dimethyl-1-[(trans-4-methylcyclohexyl)carbonyl]hydrazinyl}thiophene-2-carboxylic acid, RNA-directed RNA polymerase | | Authors: | Appleby, T.C, Canales, E, Watkins, W.J. | | Deposit date: | 2012-04-13 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Tri-substituted acylhydrazines as tertiary amide bioisosteres: HCV NS5B polymerase inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6NZV
 
 | | Crystal structure of HCV NS3/4A protease in complex with compound 12 | | Descriptor: | (1aR,5S,8S,9S,10R,22aR)-5-tert-butyl-N-[(1R,2R)-2-(difluoromethyl)-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclopropyl]-9-ethyl-14-methoxy-3,6-dioxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadecino[11,12-b]quinoxaline-8-carboxamide, HCV NS3/4A protease, SULFATE ION, ... | | Authors: | Appleby, T.C, Taylor, J.G. | | Deposit date: | 2019-02-14 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of the pan-genotypic hepatitis C virus NS3/4A protease inhibitor voxilaprevir (GS-9857): A component of Vosevi®.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6NZT
 
 | |
6X3Y
 
 | | Human cyclophilin A bound to a series of acylcic and macrocyclic inhibitors | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, tert-butyl [(2S)-1-{[(3S,17S)-2,16-dioxo-10,15-dioxa-1,21-diazatricyclo[15.3.1.1~5,9~]docosa-5(22),6,8-trien-3-yl]amino}-3-methyl-1-oxobutan-2-yl]carbamate | | Authors: | Appleby, T.C, Paulsen, J.L, Schmitz, U, Shivakumar, D. | | Deposit date: | 2020-05-21 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evaluation of Free Energy Calculations for the Prioritization of Macrocycle Synthesis.
J.Chem.Inf.Model., 60, 2020
|
|
