5ZMA
 
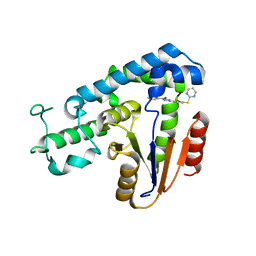 | | Structural basis for an allosteric Eya2 phosphatase inhibitor | | Descriptor: | 3-fluoro-N'-[(E)-{5-[(pyrimidin-2-yl)sulfanyl]furan-2-yl}methylidene]benzohydrazide, Eyes absent homolog 2 | | Authors: | Anantharajan, J, Jansson, A.E, Kang, C. | | Deposit date: | 2018-04-02 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.175 Å) | | Cite: | Structural and Functional Analyses of an Allosteric EYA2 Phosphatase Inhibitor That Has On-Target Effects in Human Lung Cancer Cells.
Mol.Cancer Ther., 18, 2019
|
|
2YQ8
 
 | |
8JVE
 
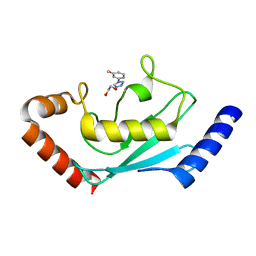 | | Identification and characterization of inhibitors covalently modifying catalytic cysteine of UBE2T and blocking ubiquitin transfer | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-methoxyphenyl)-1,2,3,4-tetrazole, Ubiquitin-conjugating enzyme E2 T | | Authors: | Anantharajan, J, Baburajendran, N. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Identification and characterization of inhibitors covalently modifying catalytic cysteine of UBE2T and blocking ubiquitin transfer.
Biochem.Biophys.Res.Commun., 689, 2023
|
|
8JVL
 
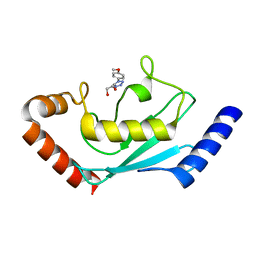 | | Identification and characterization of inhibitors covalently modifying catalytic cysteine of UBE2T and blocking ubiquitin transfer | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-methoxyphenyl)-1,2,3,4-tetrazole, Ubiquitin-conjugating enzyme E2 T | | Authors: | Anantharajan, J, Baburajendran, N. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Identification and characterization of inhibitors covalently modifying catalytic cysteine of UBE2T and blocking ubiquitin transfer.
Biochem.Biophys.Res.Commun., 689, 2023
|
|
7F8G
 
 | |
7F8H
 
 | |
4BTB
 
 | |
4BT8
 
 | |
4BTA
 
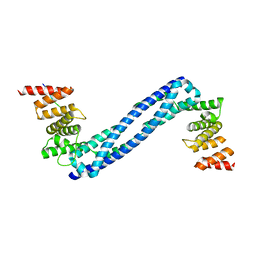 | | CRYSTAL STRUCTURE OF THE PEPTIDE(PRO-PRO-GLY)3 BOUND COMPLEX OF N- TERMINAL DOMAIN AND PEPTIDE SUBSTRATE BINDING DOMAIN OF PROLYL-4 HYDROXYLASE (RESIDUES 1-244) TYPE I FROM HUMAN | | Descriptor: | PROLINE RICH PEPTIDE, PROLYL 4-HYDROXYLASE SUBUNIT ALPHA-1 | | Authors: | Anantharajan, J, Koski, M.K, Pekkala, M, Wierenga, R.K. | | Deposit date: | 2013-06-14 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The Structural Motifs for Substrate Binding and Dimerization of the Alpha Subunit of Collagen Prolyl 4-Hydroxylase
Structure, 21, 2013
|
|
4BT9
 
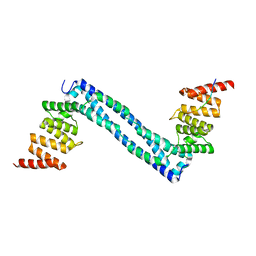 | |
9LYF
 
 | |
7CC2
 
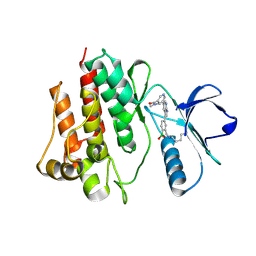 | |
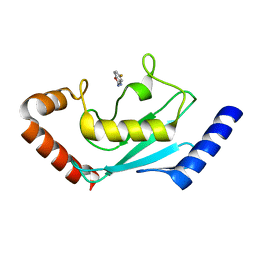
8JVD
 
 | |
8JUC
 
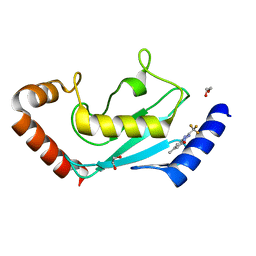 | | Identification of small-molecule binding sites of a ubiquitin-conjugating enzyme-UBE2T through fragment-based screening | | Descriptor: | 1,2-ETHANEDIOL, 7-methyl-2-(trifluoromethyl)-3~{H}-[1,2,4]triazolo[1,5-a]pyridin-5-one, Ubiquitin-conjugating enzyme E2 T | | Authors: | Anantharajan, J, Baburajendran, N. | | Deposit date: | 2023-06-26 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Identification of small-molecule binding sites of a ubiquitin-conjugating enzyme-UBE2T through fragment-based screening.
Protein Sci., 33, 2024
|
|
7DXL
 
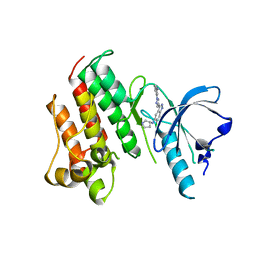 | |
7DT2
 
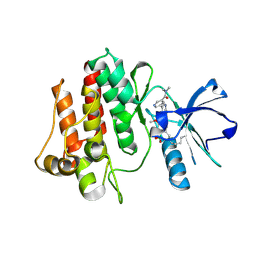 | |
6EVM
 
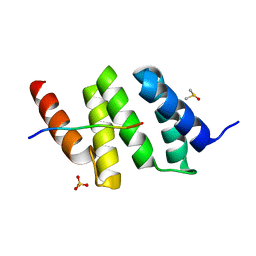 | | Crystal structure of a Pro-9 complexed peptide-substrate-binding domain of human type II collagen prolyl 4-hydroxylase | | Descriptor: | DIMETHYL SULFOXIDE, Pro-9, Prolyl 4-hydroxylase subunit alpha-2, ... | | Authors: | Murthy, A.V, Sulu, R, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural enzymology binding studies of the peptide-substrate-binding domain of human collagen prolyl 4-hydroxylase (type-II): High affinity peptides have a PxGP sequence motif.
Protein Sci., 27, 2018
|
|
6EVN
 
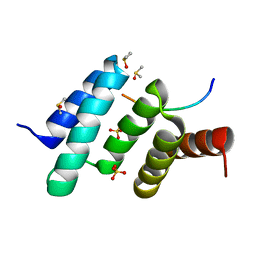 | | Crystal structure of peptide-substrate-binding domain of human type II collagen prolyl 4-hydroxylase complex with Pro-Pro-Gly-Pro-Ala-Gly-Pro-Pro-Gly. | | Descriptor: | DIMETHYL SULFOXIDE, PRO-PRO-GLY-PRO-ALA-GLY-PRO-PRO-GLY, Prolyl 4-hydroxylase subunit alpha-2, ... | | Authors: | Murthy, A.V, Sulu, R, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural enzymology binding studies of the peptide-substrate-binding domain of human collagen prolyl 4-hydroxylase (type-II): High affinity peptides have a PxGP sequence motif.
Protein Sci., 27, 2018
|
|
6EVL
 
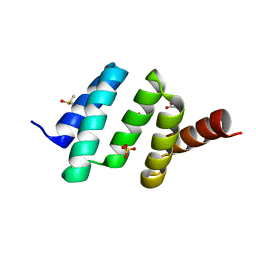 | | Crystal structure of an unlignaded peptide-substrate-binding domain of human type II collagen prolyl 4-hydroxylase | | Descriptor: | DIMETHYL SULFOXIDE, GLYCINE, Prolyl 4-hydroxylase subunit alpha-2, ... | | Authors: | Murthy, A.V, Sulu, R, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural enzymology binding studies of the peptide-substrate-binding domain of human collagen prolyl 4-hydroxylase (type-II): High affinity peptides have a PxGP sequence motif.
Protein Sci., 27, 2018
|
|
6EVP
 
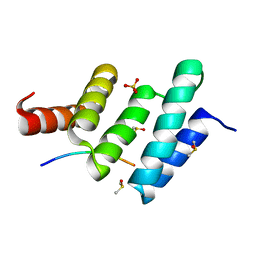 | | Crystal structure the peptide-substrate-binding domain of human type II collagen prolyl 4-hydroxylase complexed with Pro-Pro-Gly-Pro-Glu-Gly-Pro-Pro-Gly. | | Descriptor: | DIMETHYL SULFOXIDE, PRO-PRO-GLY-PRO-GLU-GLY-PRO-PRO-GLY, Prolyl 4-hydroxylase subunit alpha-2, ... | | Authors: | Murthy, A.V, Sulu, R, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural enzymology binding studies of the peptide-substrate-binding domain of human collagen prolyl 4-hydroxylase (type-II): High affinity peptides have a PxGP sequence motif.
Protein Sci., 27, 2018
|
|
6EVO
 
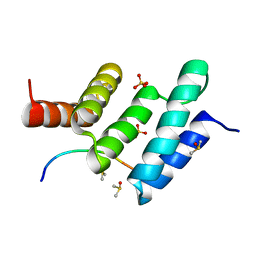 | | Crystal structure the peptide-substrate-binding domain of human type II collagen prolyl 4-hydroxylase complexed with Pro-Pro-Gly-Pro-Arg-Gly-Pro-Pro-Gly. | | Descriptor: | DIMETHYL SULFOXIDE, PRO-PRO-GLY-PRO-ARG-GLY-PRO-PRO-GLY, Prolyl 4-hydroxylase subunit alpha-2, ... | | Authors: | Murthy, A.V, Sulu, R, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural enzymology binding studies of the peptide-substrate-binding domain of human collagen prolyl 4-hydroxylase (type-II): High affinity peptides have a PxGP sequence motif.
Protein Sci., 27, 2018
|
|
