1IK1
 
 | | Solution Structure of an RNA Hairpin from HRV-14 | | Descriptor: | 5'-R(*GP*GP*UP*AP*CP*UP*AP*UP*GP*UP*AP*CP*CP*A)-3' | | Authors: | Huang, H, Alexandrov, A, Chen, X, Barnes III, T.W, Zhang, H, Dutta, K, Pascal, S.M. | | Deposit date: | 2001-05-01 | | Release date: | 2001-07-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of an RNA hairpin from HRV-14.
Biochemistry, 40, 2001
|
|
1QOJ
 
 | | Crystal Structure of E.coli UvrB C-terminal domain, and a model for UvrB-UvrC interaction. | | Descriptor: | UVRB | | Authors: | Sohi, M, Alexandrovich, A, Moolenaar, G, Visse, R, Goosen, N, Vernede, X, Fontecilla-Camps, J, Champness, J, Sanderson, M.R. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-10 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of E.Coli Uvrb C-Terminal Domain, and a Model for Uvrb-Uvrc Interaction
FEBS Lett., 465, 2000
|
|
2GIL
 
 | | Structure of the extremely slow GTPase Rab6A in the GTP bound form at 1.8 resolution | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-6A | | Authors: | Bergbrede, T, Pylypenko, O, Rak, A, Alexandrov, K. | | Deposit date: | 2006-03-29 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of the extremely slow GTPase Rab6A in the GTP bound form at 1.8 resolution
J.STRUCT.BIOL., 152, 2005
|
|
4L9X
 
 | | Triazine hydrolase from Arthobacter aurescens modified for maximum expression in E.coli | | Descriptor: | ACETATE ION, Triazine hydrolase | | Authors: | Jackson, C.J, Coppin, C.W, Alexandrov, A, Wilding, M, Liu, J.-W, Ubels, J, Paks, M, Carr, P.D, Newman, J, Russell, R.J, Field, M, Weik, M, Oakeshott, J.G, Scott, C. | | Deposit date: | 2013-06-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 300-Fold increase in production of the Zn2+-dependent dechlorinase TrzN in soluble form via apoenzyme stabilization.
Appl.Environ.Microbiol., 80, 2014
|
|
4LH8
 
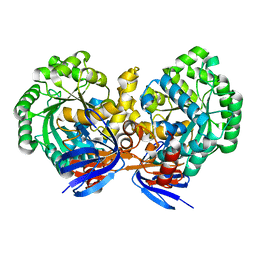 | | Triazine hydrolase from Arthobacter aurescens modified for maximum expression in E.coli | | Descriptor: | Triazine hydrolase, ZINC ION | | Authors: | Jackson, C.J, Coppin, C.W, Alexandrov, A, Wilding, M, Liu, J.-W, Ubels, J, Paks, M, Carr, P.D, Newman, J, Russell, R.J, Field, M, Weik, M, Oakeshott, J.G, Scott, C. | | Deposit date: | 2013-07-01 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 300-Fold increase in production of the Zn2+-dependent dechlorinase TrzN in soluble form via apoenzyme stabilization.
Appl.Environ.Microbiol., 80, 2014
|
|
8OSD
 
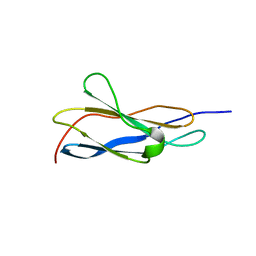 | | Crystal structure of the titin domain Fn3-49 | | Descriptor: | Titin | | Authors: | Nikoopour, R, Rees, M, Gautel, M. | | Deposit date: | 2023-04-18 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure determination and analysis of titin A-band fibronectin type III domains provides insights for disease-linked variants and protein oligomerisation.
J.Struct.Biol., 215, 2023
|
|
8OQ9
 
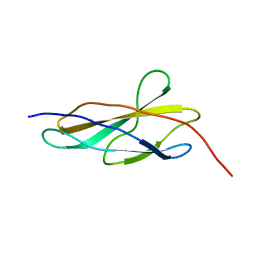 | | Crystal structure of the titin domain Fn3-56 | | Descriptor: | CHLORIDE ION, Titin, ZINC ION | | Authors: | Rees, M, Gautel, M. | | Deposit date: | 2023-04-11 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure determination and analysis of titin A-band fibronectin type III domains provides insights for disease-linked variants and protein oligomerisation.
J.Struct.Biol., 215, 2023
|
|
8ORL
 
 | |
8OTY
 
 | | Crystal structure of the titin domain Fn3-90 | | Descriptor: | Titin | | Authors: | Nikoopour, R, Rees, M, Gautel, M. | | Deposit date: | 2023-04-21 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure determination and analysis of titin A-band fibronectin type III domains provides insights for disease-linked variants and protein oligomerisation.
J.Struct.Biol., 215, 2023
|
|
8OIY
 
 | |
8OMW
 
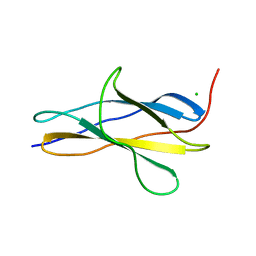 | | Crystal structure of the titin domain Fn3-20 | | Descriptor: | CHLORIDE ION, Titin | | Authors: | Rees, M, Gautel, M. | | Deposit date: | 2023-03-31 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structure determination and analysis of titin A-band fibronectin type III domains provides insights for disease-linked variants and protein oligomerisation.
J.Struct.Biol., 215, 2023
|
|
8OS3
 
 | | Crystal structure of the titin domain Fn3-11 | | Descriptor: | Titin | | Authors: | Nikoopour, R, Rees, M, Gautel, M. | | Deposit date: | 2023-04-17 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure determination and analysis of titin A-band fibronectin type III domains provides insights for disease-linked variants and protein oligomerisation.
J.Struct.Biol., 215, 2023
|
|
8OT5
 
 | | Crystal structure of the titin domain Fn3-85 | | Descriptor: | CHLORIDE ION, SODIUM ION, Titin | | Authors: | Nikoopour, R, Rees, M, Gautel, M. | | Deposit date: | 2023-04-20 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure determination and analysis of titin A-band fibronectin type III domains provides insights for disease-linked variants and protein oligomerisation.
J.Struct.Biol., 215, 2023
|
|
