7A5O
 
 | | Human MUC2 AAs 21-1397 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Javitt, G, Khmelnitsky, L, Albert, L, Elad, N, Ilani, T, Diskin, R, Fass, D. | | Deposit date: | 2020-08-21 | | Release date: | 2020-10-21 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Assembly Mechanism of Mucin and von Willebrand Factor Polymers.
Cell, 183, 2020
|
|
7PKJ
 
 | | Streptococcus pyogenes apo GapN | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, Putative NADP-dependent glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Schindelin, H, Albert, L. | | Deposit date: | 2021-08-25 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | The Non-phosphorylating Glyceraldehyde-3-Phosphate Dehydrogenase GapN Is a Potential New Drug Target in Streptococcus pyogenes.
Front Microbiol, 13, 2022
|
|
7PKC
 
 | | Streptococcus pyogenes Apo-GapN C284S variant | | Descriptor: | GLYCEROL, Putative NADP-dependent glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | Authors: | Schindelin, H, Albert, L. | | Deposit date: | 2021-08-25 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Non-phosphorylating Glyceraldehyde-3-Phosphate Dehydrogenase GapN Is a Potential New Drug Target in Streptococcus pyogenes.
Front Microbiol, 13, 2022
|
|
6TM2
 
 | | Human MUC2 AAs 21-1397 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Javitt, G, Khmelnitsky, L, Albert, L, Elad, N, Ilani, T, Diskin, R, Fass, D. | | Deposit date: | 2019-12-03 | | Release date: | 2020-10-21 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Assembly Mechanism of Mucin and von Willebrand Factor Polymers.
Cell, 183, 2020
|
|
7AXU
 
 | |
7AXQ
 
 | |
7AXX
 
 | |
7AXP
 
 | |
7AXS
 
 | |
6IAM
 
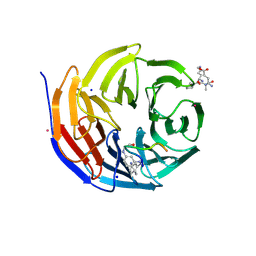 | |
5M25
 
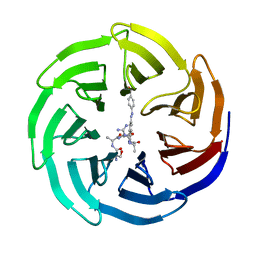 | | Modulation of MLL1 Methyltransferase Activity | | Descriptor: | (2~{S})-2-[[(2~{S})-2-[[(2~{S})-2-azanyl-3-oxidanyl-propanoyl]amino]propanoyl]amino]-5-carbamimidamido-~{N}-[(2~{S})-1-[[4-[(~{E})-[4-(hydroxymethyl)phenyl]diazenyl]phenyl]methylamino]-1-oxidanylidene-propan-2-yl]pentanamide, WD repeat-containing protein 5 | | Authors: | Srinivasan, V. | | Deposit date: | 2016-10-11 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Controlled inhibition of methyltransferases using photoswitchable peptidomimetics: towards an epigenetic regulation of leukemia.
Chem Sci, 8, 2017
|
|
5M23
 
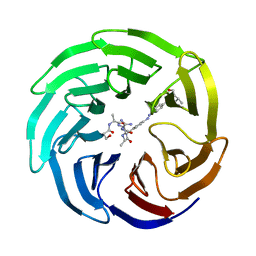 | | Modulation of MLL1 Methyltransferase Activity | | Descriptor: | 4-[(~{E})-[4-[[[(2~{S})-2-[[(2~{S})-2-[[(2~{S})-2-[[(2~{S})-2-azanyl-3-oxidanyl-propanoyl]amino]propanoyl]amino]-5-carbamimidamido-pentanoyl]amino]propanoyl]amino]methyl]phenyl]diazenyl]-~{N}-[(2~{S})-3-methyl-1-oxidanylidene-butan-2-yl]benzamide, WD repeat-containing protein 5 | | Authors: | Srinivasan, V. | | Deposit date: | 2016-10-11 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Controlled inhibition of methyltransferases using photoswitchable peptidomimetics: towards an epigenetic regulation of leukemia.
Chem Sci, 8, 2017
|
|
8C8T
 
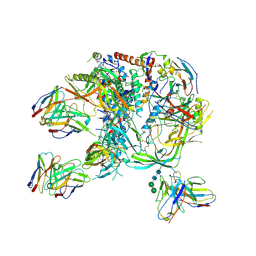 | | cryo-EM structure of BG505 SOSIP.664 HIV-1 Env trimer in complex with bNAbs EPTC112 and 3BNC117 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Baquero, E, Molinos-Albert, L, Mouquet, H. | | Deposit date: | 2023-01-21 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Anti-V1/V3-glycan broadly HIV-1 neutralizing antibodies in a post-treatment controller.
Cell Host Microbe, 31, 2023
|
|
6RBF
 
 | | Mucin 2 D3 domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Javitt, G, Fass, D. | | Deposit date: | 2019-04-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Intestinal Gel-Forming Mucins Polymerize by Disulfide-Mediated Dimerization of D3 Domains.
J.Mol.Biol., 431, 2019
|
|
6TM6
 
 | | MUC2 CysD1 domain | | Descriptor: | CALCIUM ION, Mucin-2 | | Authors: | Khmelnitsky, L, Fass, D. | | Deposit date: | 2019-12-03 | | Release date: | 2020-02-19 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Assembly Mechanism of Mucin and von Willebrand Factor Polymers.
Cell, 183, 2020
|
|
8C5H
 
 | |
