6FHE
 
 | | Highly active enzymes by automated modular backbone assembly and sequence design | | Descriptor: | Synthetic construct | | Authors: | Lapidot, G, Khersonsky, O, Lipsh, R, Dym, O, Albeck, S, Rogotner, S, Fleishman, J.S. | | Deposit date: | 2018-01-14 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Highly active enzymes by automated combinatorial backbone assembly and sequence design.
Nat Commun, 9, 2018
|
|
6FFW
 
 | | Phosphotriesterase PTE_A53_5 | | Descriptor: | (4~{S},6~{R})-2,2,6-trimethyl-1,3-dioxan-4-ol, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-01-09 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.495 Å) | | Cite: | Phosphotriesterase
PTE_A53_5
To Be Published
|
|
6FHF
 
 | | Highly active enzymes by automated modular backbone assembly and sequence design | | Descriptor: | Design, SODIUM ION | | Authors: | Lapidot, G, Khersonsky, O, Lipsh, R, Dym, O, Albeck, S, Rogotner, S, Fleishman, J.S. | | Deposit date: | 2018-01-14 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Highly active enzymes by automated combinatorial backbone assembly and sequence design.
Nat Commun, 9, 2018
|
|
6FQE
 
 | | Phosphotriesterase PTE_A53_4 | | Descriptor: | (4~{S},6~{R})-2,2,6-trimethyl-1,3-dioxan-4-ol, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Phosphotriesterase
PTE_A53_4
To Be Published
|
|
6GC2
 
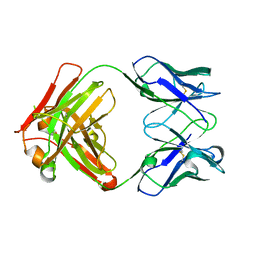 | | AbLIFT: Antibody stability and affinity optimization by computational design of the variable light-heavy chain interface | | Descriptor: | Heavy chain, Light Chain | | Authors: | Warszawski, S, Katz, A, Khmelnitsky, L, Ben Nissan, G, Javitt, G, Dym, O, Unger, T, Knop, O, Diskin, R, Albeck, S, Fass, D, Sharon, M, Fleishman, S.J. | | Deposit date: | 2018-04-17 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Optimizing antibody affinity and stability by the automated design of the variable light-heavy chain interfaces.
Plos Comput.Biol., 15, 2019
|
|
6G1J
 
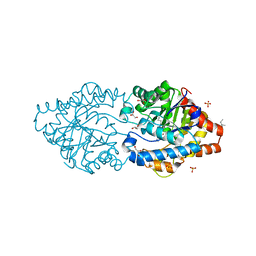 | | Phosphotriesterase PTE_C23M_1 | | Descriptor: | 1-ethyl-1-methyl-cyclohexane, FORMIC ACID, GLYCEROL, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-03-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phosphotriesterase
PTE_C23M_1
To Be Published
|
|
6FRZ
 
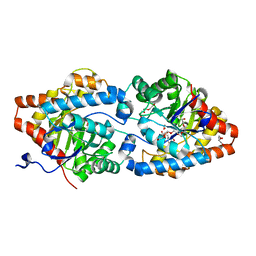 | | Phosphotriesterase PTE_A53_7 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-methylcyclohexane-1,1,3,3-tetrol, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-02-18 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Phosphotriesterase
PTE_A53_7
To Be Published
|
|
6FWE
 
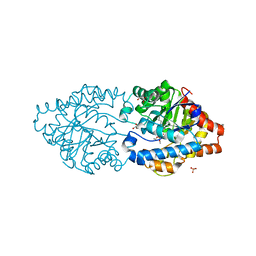 | | Phosphotriesterase PTE_C23_6 | | Descriptor: | 1-[methoxy(methyl)phosphoryl]oxyethane, 1-ethyl-1-methyl-cyclohexane, FORMIC ACID, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-03-06 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.774 Å) | | Cite: | Phosphotriesterase
PTE_C23_6
To Be Published
|
|
6FU6
 
 | | Phosphotriesterase PTE_C23_2 | | Descriptor: | FORMIC ACID, POLYACRYLIC ACID, Parathion hydrolase, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Phosphotriesterase
PTE_A53_4
To Be Published
|
|
6G3M
 
 | | Phosphotriesterase PTE_C23M_4 | | Descriptor: | 1-ethyl-1-methyl-cyclohexane, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-03-26 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Phosphotriesterase
PTE_C23M_4
To Be Published
|
|
3RON
 
 | | Crystal Structure and Hemolytic Activity of the Cyt1Aa Toxin from Bacillus thuringiensis subsp. israelensis | | Descriptor: | Type-1Aa cytolytic delta-endotoxin | | Authors: | Cohen, S, Albeck, S, Ben-Dov, E, Cahan, R, Firer, M, Zaritsky, A, Dym, O, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-04-26 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Cyt1Aa Toxin: Crystal Structure Reveals Implications for Its Membrane-Perforating Function.
J.Mol.Biol., 413, 2011
|
|
3HQJ
 
 | | Structure-function analysis of Mycobacterium tuberculosis acyl carrier protein synthase (AcpS). | | Descriptor: | COENZYME A, Holo-[acyl-carrier-protein] synthase, MAGNESIUM ION | | Authors: | Dym, O, Albeck, S, Peleg, Y, Schwarz, A, Shakked, Z, Burstein, Y, Zimhony, O, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2009-06-07 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-function analysis of the acyl carrier protein synthase (AcpS) from Mycobacterium tuberculosis.
J.Mol.Biol., 393, 2009
|
|
3UYC
 
 | | Designed protein KE59 R8_2/7A | | Descriptor: | Kemp eliminase KE59 R8_2/7A, PHOSPHATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-06 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UXD
 
 | | Designed protein KE59 R1 7/10H with dichlorobenzotriazole (DBT) | | Descriptor: | 5,7-dichloro-1H-benzotriazole, Kemp eliminase KE59 R1 7/10H, PHOSPHATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-05 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UY7
 
 | | Designed protein KE59 R1 7/10H with G130S mutation | | Descriptor: | Kemp eliminase KE59 R1 7/10H, SODIUM ION, SULFATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-06 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UXA
 
 | | Designed protein KE59 R1 7/10H | | Descriptor: | Kemp eliminase KE59 R1 7/10H, PHOSPHATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-05 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UZ5
 
 | | Designed protein KE59 R13 3/11H | | Descriptor: | 5,7-dichloro-1H-benzotriazole, Kemp eliminase KE59 R13 3/11H, PHOSPHATE ION, ... | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-07 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UZJ
 
 | | Designed protein KE59 R13 3/11H with benzotriazole | | Descriptor: | 1H-benzotriazole, Kemp eliminase KE59 R13 3/11H, PHOSPHATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-07 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UY8
 
 | | Designed protein KE59 R5_11/5F | | Descriptor: | Kemp eliminase KE59 R5_11/5F, SULFATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-06 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6ER6
 
 | | Crystal structure of a computationally designed colicin endonuclease and immunity pair colEdes7/Imdes7 | | Descriptor: | Endonuclease colEdes7, immunity Imdes7 | | Authors: | Netzer, R, Listov, D, Dym, O, Albeck, S, Knop, O, Fleishman, S.J. | | Deposit date: | 2017-10-17 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ultrahigh specificity in a network of computationally designed protein-interaction pairs.
Nat Commun, 9, 2018
|
|
6ERE
 
 | | Crystal structure of a computationally designed colicin endonuclease and immunity pair colEdes3/Imdes3 | | Descriptor: | Immunity, PHOSPHATE ION, colicin | | Authors: | Netzer, R, Listov, D, Dym, O, Albeck, S, Knop, O, Fleishman, S.J. | | Deposit date: | 2017-10-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Ultrahigh specificity in a network of computationally designed protein-interaction pairs.
Nat Commun, 9, 2018
|
|
3Q2D
 
 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution | | Descriptor: | 5-nitro-1H-benzotriazole, Deoxyribose phosphate aldolase | | Authors: | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Murphy, P, Dym, O, Albeck, S, Kiss, G, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2010-12-20 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
6QJZ
 
 | |
4U4M
 
 | | Crystal structure of 0.5M urea unfolded YagE, a KDG aldolase protein in complex with Pyruvate | | Descriptor: | 1,2-ETHANEDIOL, PYRUVIC ACID, UREA, ... | | Authors: | Manoj Kumar, P, Bhaskar, V, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Crystal structure of 0.5M urea unfolded YagE, a KDG aldolase protein in complex with Pyruvate
To be published
|
|
6ZBS
 
 | | Beta ODAP Synthetase (BOS) | | Descriptor: | Beta ODAP Synthetase (BOS), DI(HYDROXYETHYL)ETHER, MAGNESIUM ION | | Authors: | Dym, O. | | Deposit date: | 2020-06-09 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification and characterization of the key enzyme in the biosynthesis of the neurotoxin beta-ODAP in grass pea.
J.Biol.Chem., 298, 2022
|
|
