5H1A
 
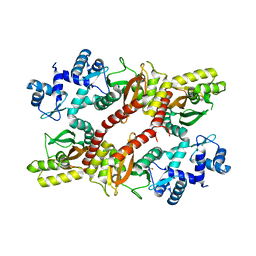 | | Crystal structure of an IclR homolog from Microbacterium sp. strain HM58-2 | | Descriptor: | IclR transcription factor homolog, PHOSPHATE ION | | Authors: | Akiyama, T, Yamada, Y, Takaya, N, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an IclR homologue from Microbacterium sp. strain HM58-2.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5H6S
 
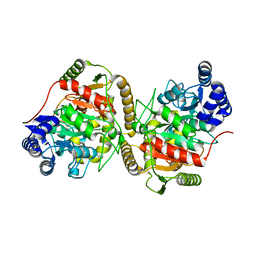 | | Crystal structure of Hydrazidase S179A mutant complexed with a substrate | | Descriptor: | 4-oxidanylbenzohydrazide, Amidase | | Authors: | Akiyama, T, Ishii, M, Takuwa, A, Oinuma, K, Sasaki, Y, Takaya, N, Yajima, S. | | Deposit date: | 2016-11-15 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the substrate recognition of hydrazidase isolated from Microbacterium sp. strain HM58-2, which catalyzes acylhydrazide compounds as its sole carbon source
Biochem. Biophys. Res. Commun., 482, 2017
|
|
5H6T
 
 | | Crystal structure of Hydrazidase from Microbacterium sp. strain HM58-2 | | Descriptor: | Amidase | | Authors: | Akiyama, T, Ishii, M, Takuwa, A, Oinuma, K, Sasaki, Y, Takaya, N, Yajima, S. | | Deposit date: | 2016-11-15 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of the substrate recognition of hydrazidase isolated from Microbacterium sp. strain HM58-2, which catalyzes acylhydrazide compounds as its sole carbon source
Biochem. Biophys. Res. Commun., 482, 2017
|
|
7CUO
 
 | | IclR transcription factor complexed with 4-hydroxybenzoic acid from Microbacterium hydrocarbonoxydans | | Descriptor: | P-HYDROXYBENZOIC ACID, SULFATE ION, Transcription factor | | Authors: | Akiyama, T, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-08-23 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the conformational changes in Microbacterium hydrocarbonoxydans IclR transcription factor homolog due to ligand binding.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
7DQB
 
 | | Crystal structure of an IclR homolog complexed with 4-hydroxybenzoate from Microbacterium hydrocarbonoxydans in P212121 form | | Descriptor: | IclR homolog, P-HYDROXYBENZOIC ACID | | Authors: | Akiyama, T, Sasaki, Y, Ito, S, Yajima, S. | | Deposit date: | 2020-12-23 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural basis of the conformational changes in Microbacterium hydrocarbonoxydans IclR transcription factor homolog due to ligand binding.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
5B2H
 
 | | Crystal structure of HA33 from Clostridium botulinum serotype C strain Yoichi | | Descriptor: | HA-33, TRIETHYLENE GLYCOL | | Authors: | Akiyama, T, Hayashi, S, Matsumoto, T, Hasegawa, K, Yamano, A, Suzuki, T, Niwa, K, Watanabe, T, Sagane, Y, Yajima, S. | | Deposit date: | 2016-01-15 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational divergence in the HA-33/HA-17 trimer of serotype C and D botulinum toxin complex
Biochem.Biophys.Res.Commun., 476, 2016
|
|
5YGV
 
 | | Crystal structure of the abscisic acid receptor PYR1 in complex with an antagonist | | Descriptor: | (2Z,4E)-3-methyl-5-[(1S,4S)-2,6,6-trimethyl-4-[3-(4-methylphenyl)prop-2-ynoxy]-1-oxidanyl-cyclohex-2-en-1-yl]penta-2,4-dienoic acid, Abscisic acid receptor PYR1 | | Authors: | Akiyama, T, Sue, M, Takeuchi, J, Mimura, N, Okamoto, M, Monda, K, Iba, K, Ohnishi, T, Todoroki, Y, Yajima, S. | | Deposit date: | 2017-09-27 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Chemical Design of Abscisic Acid Antagonists That Block PYL-PP2C Receptor Interactions.
ACS Chem. Biol., 13, 2018
|
|
3WG8
 
 | | Crystal structure of the abscisic acid receptor PYR1 in complex with an antagonist AS6 | | Descriptor: | (2Z,4E)-5-[(1S)-3-(hexylsulfanyl)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYR1 | | Authors: | Akiyama, T, Sue, M, Takeuchi, J, Okamoto, M, Muto, T, Endo, A, Nambara, E, Hirai, N, Ohnishi, T, Cutler, S.R, Todoroki, Y, Yajima, S. | | Deposit date: | 2013-07-31 | | Release date: | 2014-05-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Designed abscisic acid analogs as antagonists of PYL-PP2C receptor interactions
Nat.Chem.Biol., 10, 2014
|
|
2J4X
 
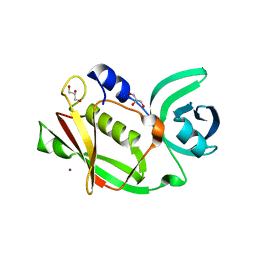 | | Streptococcus dysgalactiae-derived mitogen (SDM) | | Descriptor: | GLYCEROL, MITOGEN, ZINC ION | | Authors: | Saarinen, S, Kato, H, Uchiyama, T, Miyoshi-Akiyama, T, Papageorgiou, A.C. | | Deposit date: | 2006-09-07 | | Release date: | 2007-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Streptococcus Dysgalactiae-Derived Mitogen Reveals a Zinc-Binding Site and Alterations in Tcr Binding.
J.Mol.Biol., 373, 2007
|
|
3GNR
 
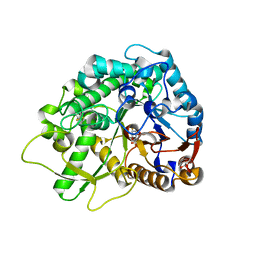 | | Crystal Structure of a Rice Os3BGlu6 Beta-Glucosidase with covalently bound 2-deoxy-2-fluoroglucoside to the catalytic nucleophile E396 | | Descriptor: | 2-deoxy-2-fluoro-alpha-D-glucopyranose, GLYCEROL, Os03g0212800 protein | | Authors: | Seshadri, S, Akiyama, T, Opassiri, R, Kuaprasert, B, Cairns, J.R.K. | | Deposit date: | 2009-03-17 | | Release date: | 2009-07-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and enzymatic characterization of Os3BGlu6, a rice beta-glucosidase hydrolyzing hydrophobic glycosides and (1->3)- and (1->2)-linked disaccharides.
Plant Physiol., 151, 2009
|
|
6LU4
 
 | | Crystal structure of the substrate binding protein from Microbacterium hydrocarbonoxydans complexed with propylparaben | | Descriptor: | Substrate binding protein, propyl 4-hydroxybenzoate | | Authors: | Shimamura, K, Akiyama, T, Yokoyama, K, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-01-25 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of substrate recognition by the substrate binding protein (SBP) of a hydrazide transporter, obtained from Microbacterium hydrocarbonoxydans.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
3QHE
 
 | | Crystal structure of the complex between the armadillo repeat domain of adenomatous polyposis coli and the tyrosine-rich domain of Sam68 | | Descriptor: | Adenomatous polyposis coli protein, KH domain-containing, RNA-binding, ... | | Authors: | Morishita, E.C.J, Murayama, K, Kato-Murayama, M, Ishizuku-Katsura, Y, Tomabechi, Y, Terada, T, Handa, N, Shirouzu, M, Akiyama, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-01-25 | | Release date: | 2011-11-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the armadillo repeat domain of adenomatous polyposis coli and its complex with the tyrosine-rich domain of sam68
Structure, 19, 2011
|
|
3GNP
 
 | | Crystal Structure of a Rice Os3BGlu6 Beta-Glucosidase with Octyl-Beta-D-Thio-Glucoside | | Descriptor: | GLYCEROL, Os03g0212800 protein, octyl 1-thio-beta-D-glucopyranoside | | Authors: | Seshadri, S, Akiyama, T, Opassiri, R, Kuaprasert, B, Cairns, J.R.K. | | Deposit date: | 2009-03-17 | | Release date: | 2009-07-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and enzymatic characterization of Os3BGlu6, a rice {beta}-glucosidase hydrolyzing hydrophobic glycosides and (1->3)- and (1->2)-linked disaccharides.
Plant Physiol., 2009
|
|
3GNO
 
 | | Crystal Structure of a Rice Os3BGlu6 Beta-Glucosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Os03g0212800 protein | | Authors: | Seshadri, S, Akiyama, T, Opassiri, R, Kuaprasert, B, Cairns, J.R.K. | | Deposit date: | 2009-03-17 | | Release date: | 2009-07-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural and enzymatic characterization of Os3BGlu6, a rice {beta}-glucosidase hydrolyzing hydrophobic glycosides and (1->3)- and (1->2)-linked disaccharides.
Plant Physiol., 2009
|
|
6KFQ
 
 | | Crystal structure of thermophilic rhodopsin from Rubrobacter xylanophilus | | Descriptor: | RETINAL, Rhodopsin, SULFATE ION, ... | | Authors: | Suzuki, K, Akiyama, T, Hayashi, T, Yasuda, S, Kanehara, K, Kojima, K, Tanabe, M, Kato, R, Senda, T, Sudo, Y, Kinoshita, M, Murata, T. | | Deposit date: | 2019-07-08 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | How Does a Microbial Rhodopsin RxR Realize Its Exceptionally High Thermostability with the Proton-Pumping Function Being Retained?
J.Phys.Chem.B, 124, 2020
|
|
6LU2
 
 | | Crystal structure of a substrate binding protein from Microbacterium hydrocarbonoxydans | | Descriptor: | Substrate binding protein | | Authors: | Shimamura, K, Akiyama, T, Yokoyama, K, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-01-25 | | Release date: | 2020-03-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of substrate recognition by the substrate binding protein (SBP) of a hydrazide transporter, obtained from Microbacterium hydrocarbonoxydans.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
6LU3
 
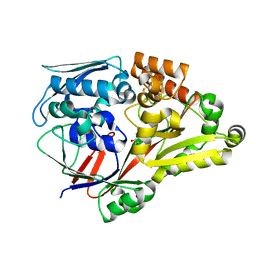 | | Crystal structure of a substrate binding protein from Microbacterium hydrocarbonoxydans complexed with 4-hydroxybenzoate hydrazide | | Descriptor: | 4-oxidanylbenzohydrazide, Substrate binding protein | | Authors: | Shimamura, K, Akiyama, T, Yokoyama, K, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-01-25 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of substrate recognition by the substrate binding protein (SBP) of a hydrazide transporter, obtained from Microbacterium hydrocarbonoxydans.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
4TRW
 
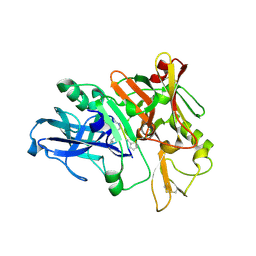 | | Structure of BACE1 complex with a syn-HEA-type inhibitor | | Descriptor: | Beta-secretase 1, L-alpha-glutamyl-L-isoleucyl-N-[(2R,3S)-1-{[(1S)-1-carboxybutyl]amino}-2-hydroxy-5-methylhexan-3-yl]-3-thiophen-2-yl-L-alaninamide | | Authors: | Akaji, K, Teruya, K, Akiyama, T, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2014-06-18 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Evaluation of transition-state mimics in a superior BACE1 cleavage sequence as peptide-mimetic BACE1 inhibitors
Bioorg.Med.Chem., 23, 2015
|
|
4TRZ
 
 | | Structure of BACE1 complex with 2-thiophenyl HEA-type inhibitor | | Descriptor: | 2-thiophenyl HEA-type inhibitor, Beta-secretase 1 | | Authors: | Akaji, K, Teruya, K, Akiyama, T, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2014-06-18 | | Release date: | 2015-07-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Evaluation of transition-state mimics in a superior BACE1 cleavage sequence as peptide-mimetic BACE1 inhibitors
Bioorg.Med.Chem., 23, 2015
|
|
4TRY
 
 | | Structure of BACE1 complex with a HEA-type inhibitor | | Descriptor: | Beta-secretase 1, GLU-ILE-TIH-THC-NVA | | Authors: | Akaji, K, Teruya, K, Akiyama, T, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2014-06-18 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of BACE1 complex with an anti-HMC-type inhibitor
to be published
|
|
3W9Y
 
 | | Crystal structure of the human DLG1 guanylate kinase domain | | Descriptor: | Disks large homolog 1 | | Authors: | Mori, S, Tezuka, Y, Arakawa, A, Handa, N, Shirouzu, M, Akiyama, T, Yokoyama, S. | | Deposit date: | 2013-04-18 | | Release date: | 2013-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the guanylate kinase domain from discs large homolog 1 (DLG1/SAP97)
Biochem.Biophys.Res.Commun., 435, 2013
|
|
2DX1
 
 | | Crystal structure of RhoGEF protein Asef | | Descriptor: | Rho guanine nucleotide exchange factor 4 | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-22 | | Release date: | 2007-01-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structure of the rac activator, Asef, reveals its autoinhibitory mechanism
J.Biol.Chem., 282, 2007
|
|
1R77
 
 | | Crystal structure of the cell wall targeting domain of peptidylglycan hydrolase ALE-1 | | Descriptor: | Cell Wall Targeting Domain of Glycylglycine Endopeptidase ALE-1 | | Authors: | Lu, J.Z, Fujiwara, T, Komatsuzawa, H, Sugai, M, Sakon, J. | | Deposit date: | 2003-10-20 | | Release date: | 2005-04-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cell Wall-targeting Domain of Glycylglycine Endopeptidase Distinguishes among Peptidoglycan Cross-bridges.
J.Biol.Chem., 281, 2006
|
|
6L9V
 
 | | Crystal structure of mouse TIFA (T9D/C36S mutant) | | Descriptor: | TRAF-interacting protein with FHA domain-containing protein A | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2019-11-11 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural analysis of TIFA: Insight into TIFA-dependent signal transduction in innate immunity.
Sci Rep, 10, 2020
|
|
6L9W
 
 | | Crystal structure of mouse TIFA (T9E/C36S mutant) | | Descriptor: | TRAF-interacting protein with FHA domain-containing protein A | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2019-11-11 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural analysis of TIFA: Insight into TIFA-dependent signal transduction in innate immunity.
Sci Rep, 10, 2020
|
|
