6ZEZ
 
 | |
6ZF5
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 1-[3-[(4-butylphenyl)sulfonyl-(2-hydroxy-2-oxoethyl)amino]phenyl]-5-cyclopropyl-pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6Z2H
 
 | |
7ABB
 
 | |
7A1A
 
 | |
7A9J
 
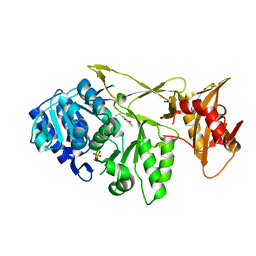 | |
7A9I
 
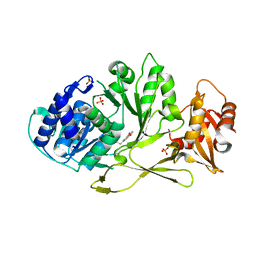 | |
8B8H
 
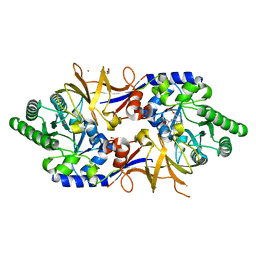 | | Structure of DCS-resistant variant D322N of alanine racemase from M. tuberculosis in complex with DCS | | Descriptor: | (~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylidene-[(4~{R})-3-oxidanylidene-1,2-oxazolidin-4-yl]azanium, 1,2-ETHANEDIOL, Alanine racemase, ... | | Authors: | de Chiara, C, Prosser, G, Ogrodowicz, R.W, de Carvalho, L.P.S. | | Deposit date: | 2022-10-04 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of the d-Cycloserine-Resistant Variant D322N of Alanine Racemase from Mycobacterium tuberculosis .
Acs Bio Med Chem Au, 3, 2023
|
|
8ASO
 
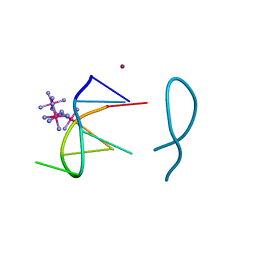 | | Nickel(II) bound to a non-canonical quadruplex | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*GP*CP*AP*TP*GP*CP*T)-3'), NICKEL (II) ION | | Authors: | Lambert, M.C, Hall, J.P. | | Deposit date: | 2022-08-19 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Identifying metal-DNA binding sites, what is the best method to get transition metals into a crystal system?
To Be Published
|
|
8ASM
 
 | | Cobalt(II) bound to a non-canonical quadruplex | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), DNA (5'-D(*GP*CP*AP*TP*GP*CP*T)-3') | | Authors: | Lambert, M.C, Hall, J.P. | | Deposit date: | 2022-08-19 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identifying metal-DNA binding sites, what is the best method to get transition metals into a crystal system?
To Be Published
|
|
8AZC
 
 | |
8AMS
 
 | | Complex of human TRIM2 RING domain, UBCH5C, and Ubiquitin | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, GLYCEROL, Polyubiquitin-C, ... | | Authors: | Perez-Borrajero, C, Kotova, I, Murciano, B, Hennig, J. | | Deposit date: | 2022-08-04 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biophysical studies of TRIM2 and TRIM3
To Be Published
|
|
8B23
 
 | |
8B22
 
 | |
8B24
 
 | | Time-resolved structure of K+-dependent Na+-PPase from Thermotoga maritima 3600-seconds post reaction initiation with Na+ | | Descriptor: | DIPHOSPHATE, K(+)-stimulated pyrophosphate-energized sodium pump, MAGNESIUM ION, ... | | Authors: | Strauss, J, Vidilaseris, K, Goldman, A. | | Deposit date: | 2022-09-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (4.53 Å) | | Cite: | Functional and structural asymmetry suggest a unifying principle for catalysis in membrane-bound pyrophosphatases.
Embo Rep., 25, 2024
|
|
8B21
 
 | | Time-resolved structure of K+-dependent Na+-PPase from Thermotoga maritima 0-60-seconds post reaction initiation with Na+ | | Descriptor: | DI(HYDROXYETHYL)ETHER, DODECYL-BETA-D-MALTOSIDE, K(+)-stimulated pyrophosphate-energized sodium pump, ... | | Authors: | Strauss, J, Vidilaseris, K, Goldman, A. | | Deposit date: | 2022-09-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Functional and structural asymmetry suggest a unifying principle for catalysis in membrane-bound pyrophosphatases.
Embo Rep., 25, 2024
|
|
8BHD
 
 | | N-terminal domain of Plasmodium berghei glutamyl-tRNA synthetase (Tbxo4 derivative crystal structure) | | Descriptor: | GLYCEROL, Glutamate--tRNA ligase, SULFATE ION, ... | | Authors: | Benas, P, Jaramillo Ponce, J.R, Legrand, P, Frugier, M, Sauter, C. | | Deposit date: | 2022-10-31 | | Release date: | 2023-01-25 | | Last modified: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Solution X-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci., 32, 2023
|
|
8C7R
 
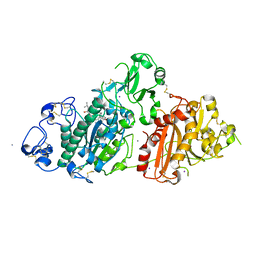 | | Crystal structure of rat autotaxin and compound MEY-003 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5,7-bis(oxidanyl)-2-(1-pentylindol-3-yl)chromen-4-one, 7alpha-hydroxycholesterol, ... | | Authors: | Eymery, M.C, McCarthy, A.A. | | Deposit date: | 2023-01-17 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Discovery of potent chromone-based autotaxin inhibitors inspired by cannabinoids.
Eur.J.Med.Chem., 263, 2023
|
|
8C4W
 
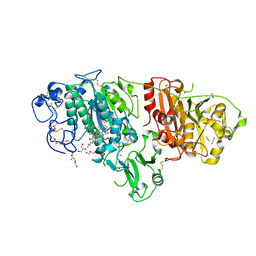 | | Crystal structure of rat autotaxin and compound MEY-002 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5,7-bis(oxidanyl)-2-[1-(phenylmethyl)indol-3-yl]chromen-4-one, 7alpha-hydroxycholesterol, ... | | Authors: | Eymery, M.C, McCarthy, A.A. | | Deposit date: | 2023-01-05 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of potent chromone-based autotaxin inhibitors inspired by cannabinoids.
Eur.J.Med.Chem., 263, 2023
|
|
8C3O
 
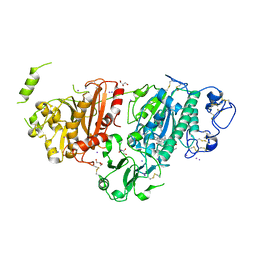 | | Crystal structure of autotaxin gamma and compound MEY-003 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5,7-bis(oxidanyl)-2-(1-pentylindol-3-yl)chromen-4-one, 7alpha-hydroxycholesterol, ... | | Authors: | Eymery, M.C, McCarthy, A.A. | | Deposit date: | 2022-12-27 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Discovery of potent chromone-based autotaxin inhibitors inspired by cannabinoids.
Eur.J.Med.Chem., 263, 2023
|
|
8C3P
 
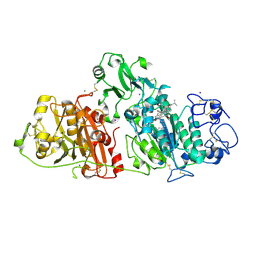 | |
8C3T
 
 | |
8CEM
 
 | | Structure of bovine native C3, re-refinement | | Descriptor: | Complement C3 alpha chain, Complement C3 beta chain, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Andersen, G.R, Fredslund, F. | | Deposit date: | 2023-02-02 | | Release date: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of bovine complement component 3 reveals the basis for thioester function.
J Mol Biol, 361, 2006
|
|
8CD8
 
 | | Ulilysin - C269A with AEBSF complex | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, CALCIUM ION, GLY-SER-SER, ... | | Authors: | Rodriguez-Banqueri, A, Eckhard, U, Gomis-Ruth, F.X. | | Deposit date: | 2023-01-30 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into latency of the metallopeptidase ulilysin (lysargiNase) and its unexpected inhibition by a sulfonyl-fluoride inhibitor of serine peptidases.
Dalton Trans, 52, 2023
|
|
8D86
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence co-crystallized with a guest small molecule, netropsin. | | Descriptor: | DNA (5'-D(A*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3'), DNA (5'-D(A*GP*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Snow, C.D. | | Deposit date: | 2022-06-07 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Modular Protein-DNA Cocrystals as Precise, Programmable Assembly Scaffolds.
Acs Nano, 17, 2023
|
|
