6ONA
 
 | |
8CTR
 
 | |
8CU9
 
 | |
3F0G
 
 | |
8DTE
 
 | |
8DTC
 
 | |
8DU1
 
 | |
8DT6
 
 | |
8DV0
 
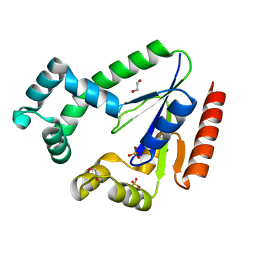 | |
3IKF
 
 | | Crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from Burkholderia pseudomallei with FOL fragment 717, imidazo[2,,1-b][1,3]thiazol-6-ylmethanol | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-08-05 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Leveraging structure determination with fragment screening for infectious disease drug targets: MECP synthase from Burkholderia pseudomallei.
J Struct Funct Genomics, 12, 2011
|
|
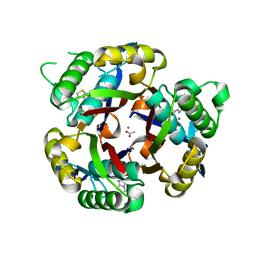
6O49
 
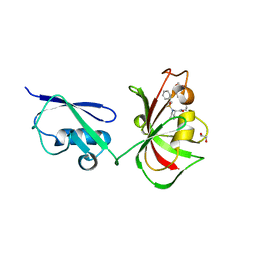 | |
6DA0
 
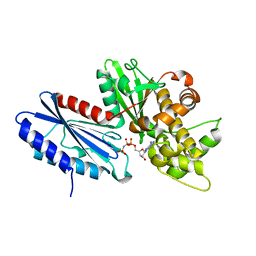 | |
3F0D
 
 | |
3F0E
 
 | |
3IEW
 
 | |
3IKE
 
 | |
3IEQ
 
 | |
3K2X
 
 | |
6ONN
 
 | |
6OSR
 
 | |
6OTE
 
 | |
4P9A
 
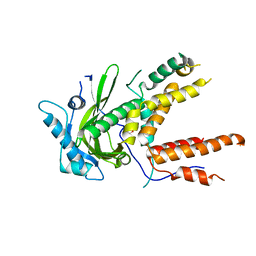 | | X-ray Crystal Structure of PA protein from Influenza strain H7N9 | | Descriptor: | GLYCEROL, Polymerase PA | | Authors: | Fairman, J.W, Moen, S.O, Clifton, M.C, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-04-02 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1999 Å) | | Cite: | Structural analysis of H1N1 and H7N9 influenza A virus PA in the absence of PB1.
Sci Rep, 4, 2014
|
|
4PFS
 
 | |
6Q09
 
 | |
6PTR
 
 | |
