8SZ5
 
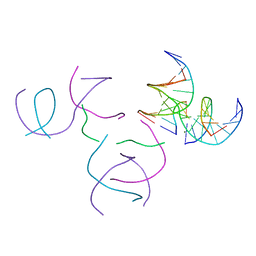 | | [2T5] Self-assembling DNA motif with 5 base pairs between junctions and P32 symmetry | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*G)-3'), DNA (5'-D(P*AP*CP*GP*AP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(P*CP*AP*CP*GP*T)-3'), ... | | Authors: | Vecchioni, S, Janowski, J, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-05-26 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
1D9X
 
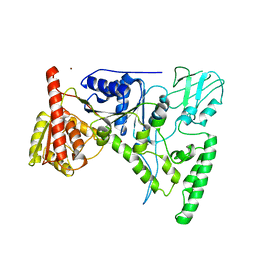 | | CRYSTAL STRUCTURE OF THE DNA REPAIR PROTEIN UVRB | | Descriptor: | EXCINUCLEASE UVRABC COMPONENT UVRB, ZINC ION | | Authors: | Theis, K, Chen, P.J, Skorvaga, M, Van Houten, B, Kisker, C. | | Deposit date: | 1999-10-30 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair.
EMBO J., 18, 1999
|
|
1D9Z
 
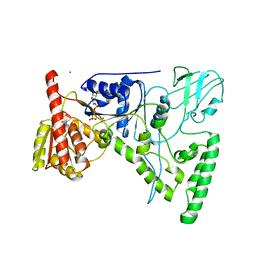 | | CRYSTAL STRUCTURE OF THE DNA REPAIR PROTEIN UVRB IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, EXCINUCLEASE UVRABC COMPONENT UVRB, MAGNESIUM ION, ... | | Authors: | Theis, K, Chen, P.J, Skorvaga, M, Van Houten, B, Kisker, C. | | Deposit date: | 1999-10-30 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair.
EMBO J., 18, 1999
|
|
7D4B
 
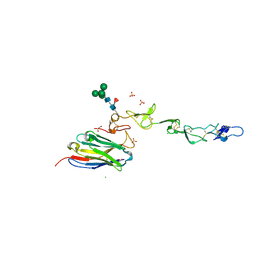 | | Crystal structure of 4-1BB in complex with a VHH | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Wang, C. | | Deposit date: | 2020-09-23 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Generation of a safe and efficacious llama single-domain antibody fragment (vHH) targeting the membrane-proximal region of 4-1BB for engineering therapeutic bispecific antibodies for cancer.
J Immunother Cancer, 9, 2021
|
|
7CZD
 
 | | Crystal structure of PD-L1 in complex with a VHH | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Programmed cell death 1 ligand 1, ... | | Authors: | Wang, C. | | Deposit date: | 2020-09-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Generation of a safe and efficacious llama single-domain antibody fragment (vHH) targeting the membrane-proximal region of 4-1BB for engineering therapeutic bispecific antibodies for cancer.
J Immunother Cancer, 9, 2021
|
|
8H08
 
 | |
8H07
 
 | |
7E2V
 
 | | Crystal structure of MaDA-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, MaDA-3 | | Authors: | Gao, L, Du, X, Fan, J, Lei, X. | | Deposit date: | 2021-02-07 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Enzymatic control of endo- and exo-stereoselective Diels-Alder reactions with broad substrate scope.
Nat Catal, 2021
|
|
5YE9
 
 | | The crystal structure of Lp-PLA2 in complex with a novel inhibitor | | Descriptor: | N-[4-[(3-cyano-4-naphthalen-2-yloxy-phenyl)sulfamoyl]phenyl]ethanamide, Platelet-activating factor acetylhydrolase, SULFATE ION | | Authors: | Liu, Q.F, Xu, Y.C. | | Deposit date: | 2017-09-15 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.876 Å) | | Cite: | Structure-Guided Discovery of Novel, Potent, and Orally Bioavailable Inhibitors of Lipoprotein-Associated Phospholipase A2.
J. Med. Chem., 60, 2017
|
|
5YE7
 
 | | The crystal structure of Lp-PLA2 in complex with a novel inhibitor | | Descriptor: | N-[4-[(4-naphthalen-2-yloxyphenyl)sulfamoyl]phenyl]ethanamide, Platelet-activating factor acetylhydrolase, SULFATE ION | | Authors: | Liu, Q.F, Xu, Y.C. | | Deposit date: | 2017-09-15 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.312 Å) | | Cite: | Structure-Guided Discovery of Novel, Potent, and Orally Bioavailable Inhibitors of Lipoprotein-Associated Phospholipase A2.
J. Med. Chem., 60, 2017
|
|
5YE8
 
 | | The crystal structure of Lp-PLA2 in complex with a novel inhibitor | | Descriptor: | N-[3,4-bis(fluoranyl)phenyl]methanesulfonamide, Platelet-activating factor acetylhydrolase | | Authors: | Liu, Q.F, Xu, Y.C. | | Deposit date: | 2017-09-15 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structure-Guided Discovery of Novel, Potent, and Orally Bioavailable Inhibitors of Lipoprotein-Associated Phospholipase A2.
J. Med. Chem., 60, 2017
|
|
5YEA
 
 | | The crystal structure of Lp-PLA2 in complex with a novel inhibitor | | Descriptor: | 4-[[4-[4-chloranyl-3-(trifluoromethyl)phenoxy]-3-cyano-phenyl]sulfamoyl]benzoic acid, Platelet-activating factor acetylhydrolase, SULFATE ION | | Authors: | Liu, Q.F, Xu, Y.C. | | Deposit date: | 2017-09-15 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Structure-Guided Discovery of Novel, Potent, and Orally Bioavailable Inhibitors of Lipoprotein-Associated Phospholipase A2.
J. Med. Chem., 60, 2017
|
|
4JYU
 
 | |
4JYV
 
 | |
8I7L
 
 | | Crystal structure of indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with a novel inhibitor | | Descriptor: | 1-[3-[(4-chloranyl-2-fluoranyl-phenyl)carbamoylamino]-4-[cyclohexyl(2-methylpropyl)amino]phenyl]pyrrole-2-carboxylic acid, Indoleamine 2,3-dioxygenase 1, THIOSULFATE | | Authors: | Li, K, Liu, W, Dong, X. | | Deposit date: | 2023-02-01 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Apo-Form Selective Inhibition of IDO for Tumor Immunotherapy.
J Immunol., 209, 2022
|
|
7X2H
 
 | |
8K4C
 
 | | Crystal structure of PDE4D complexed with ethaverine hydrochloride | | Descriptor: | 1,2-ETHANEDIOL, 1-[(3,4-diethoxyphenyl)methyl]-6,7-diethoxy-isoquinoline, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
8K4H
 
 | | Crystal structure of PDE4D complexed with benzbromarone | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, ZINC ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-07-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
7XD2
 
 | |
5I46
 
 | | Factor VIIA in complex with the inhibitor (2R,15R)-2-[(1-aminoisoquinolin-6-yl)amino]-8-fluoro-7-hydroxy-4,15,17-trimethyl-13-oxa-4,11-diazatricyclo[14.2.2.1~6,10~]henicosa-1(18),6(21),7,9,16,19-hexaene-3,12-dione | | Descriptor: | (2R,15R)-2-[(1-aminoisoquinolin-6-yl)amino]-8-fluoro-7-hydroxy-4,15,17-trimethyl-13-oxa-4,11-diazatricyclo[14.2.2.1~6,10~]henicosa-1(18),6(21),7,9,16,19-hexaene-3,12-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wei, A. | | Deposit date: | 2016-02-11 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Atropisomer Control in Macrocyclic Factor VIIa Inhibitors.
J.Med.Chem., 59, 2016
|
|
7X86
 
 | | The crystal structure of PloI4-F124L in complex with endo-4+2 adduct | | Descriptor: | (4S,4aS,6aR,8R,9R,10aS,13S,14aS,18aR,18bR,E)-9-ethyl-4,8,19-trihydroxy-10a,12,13,18a-tetramethyl-2,3,4,4a,6a,7,8,9,10,10a,13,14,18a,18b-tetradecahydro-14a,17-(metheno)benzo[b]naphtho[2,1-h][1]azacyclododecine-16,18(1H,15H)-dione, PloI4 | | Authors: | Li, M, Pan, L.F. | | Deposit date: | 2022-03-11 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | A cyclase that catalyses competing 2 + 2 and 4 + 2 cycloadditions.
Nat.Chem., 15, 2023
|
|
7X80
 
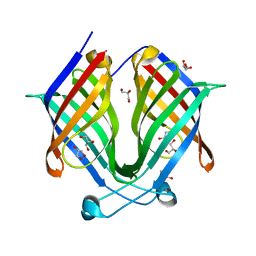 | |
7X81
 
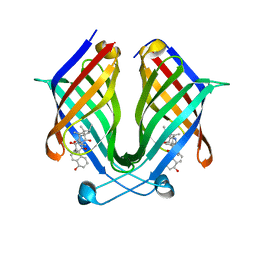 | | The crystal structure of PloI4-C16M/D46A/I137V in complex with exo-2+2 adduct | | Descriptor: | (4S,4aS,6aR,8R,9R,11E,12aR,14aS,17E,18aR,18bR)-9-ethyl-4,8,19-trihydroxy-11,12a,13,18a-tetramethyl-2,3,4,4a,6a,7,8,9,10,12a,13,14,18a,18b-tetradecahydro-14a,17-(metheno)cyclobuta[b]naphtho[2,1-j][1]azacyclotetradecine-16,18(1H,15H)-dione, PloI4 | | Authors: | Li, M, Pan, L.F. | | Deposit date: | 2022-03-10 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | A cyclase that catalyses competing 2 + 2 and 4 + 2 cycloadditions.
Nat.Chem., 15, 2023
|
|
7X7Z
 
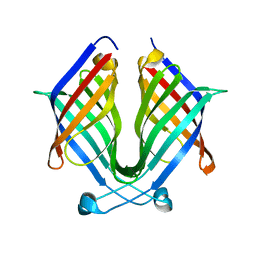 | |
8GDS
 
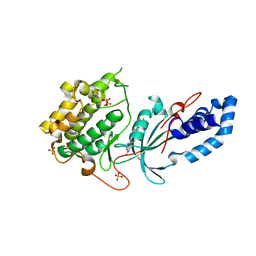 | |
