3O19
 
 | | Structure-function analysis of human L-Prostaglandin D Synthase bound with fatty acid | | Descriptor: | OLEIC ACID, PALMITIC ACID, Prostaglandin-H2 D-isomerase | | Authors: | Zhou, Y, Shaw, N, Li, Y, Zhao, Y, Zhang, R, Liu, Z.-J. | | Deposit date: | 2010-07-21 | | Release date: | 2010-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure-function analysis of human L-Prostaglandin D Synthase bound with fatty acid
To be Published
|
|
3O2Y
 
 | | Structure-function analysis of human L-Prostaglandin D Synthase bound with fatty acid | | Descriptor: | GLYCEROL, OLEIC ACID, PALMITIC ACID, ... | | Authors: | Zhou, Y, Shaw, N, Li, Y, Zhao, Y, Zhang, R, Liu, Z.-J. | | Deposit date: | 2010-07-23 | | Release date: | 2010-09-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-function analysis of human L-Prostaglandin D Synthase bound with fatty acid
To be Published
|
|
3O22
 
 | | Structure-function analysis of human L-Prostaglandin D Synthase bound with fatty acid | | Descriptor: | OLEIC ACID, PALMITIC ACID, Prostaglandin-H2 D-isomerase | | Authors: | Zhou, Y, Shaw, N, Li, Y, Zhao, Y, Zhang, R, Liu, Z.-J. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-function analysis of human L-Prostaglandin D Synthase bound with fatty acid
To be Published
|
|
8D7H
 
 | | Cryo-EM structure of human CLCF1 in complex with CRLF1 and CNTFR alpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cardiotrophin-like cytokine factor 1, ... | | Authors: | Zhou, Y, Franklin, M.C. | | Deposit date: | 2022-06-07 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the assembly of gp130 family cytokine signaling complexes.
Sci Adv, 9, 2023
|
|
8D82
 
 | |
8D7E
 
 | |
8D7R
 
 | |
8D74
 
 | |
8D6A
 
 | |
8D85
 
 | |
2K73
 
 | | Solution NMR structure of integral membrane protein DsbB | | Descriptor: | Disulfide bond formation protein B | | Authors: | Zhou, Y, Cierpicki, T, Flores Jimenez, R.H, Lukasik, S.M, Ellena, J.F, Cafiso, D.S, Kadokura, H, Beckwith, J, Bushweller, J.H. | | Deposit date: | 2008-08-01 | | Release date: | 2008-10-07 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the integral membrane enzyme DsbB: functional insights into DsbB-catalyzed disulfide bond formation.
Mol.Cell, 31, 2008
|
|
2K74
 
 | | Solution NMR structure of DsbB-ubiquinone complex | | Descriptor: | Disulfide bond formation protein B, UBIQUINONE-2 | | Authors: | Zhou, Y, Cierpicki, T, Flores Jimenez, R.H, Lukasik, S.M, Ellena, J.F, Cafiso, D.S, Kadokura, H, Beckwith, J, Bushweller, J.H. | | Deposit date: | 2008-08-01 | | Release date: | 2008-10-07 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the integral membrane enzyme DsbB: functional insights into DsbB-catalyzed disulfide bond formation.
Mol.Cell, 31, 2008
|
|
7TB0
 
 | | E. faecium MurAA in complex with fosfomycin and UNAG | | Descriptor: | CHLORIDE ION, POTASSIUM ION, SODIUM ION, ... | | Authors: | Zhou, Y, Shamoo, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Enolpyruvate transferase MurAA A149E , identified during adaptation of Enterococcus faecium to daptomycin, increases stability of MurAA-MurG interaction.
J.Biol.Chem., 299, 2023
|
|
5GUJ
 
 | |
4F4U
 
 | |
4F56
 
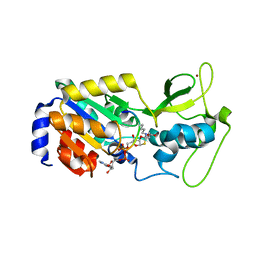 | | The bicyclic intermediate structure provides insights into the desuccinylation mechanism of SIRT5 | | Descriptor: | 3-[(2R,3aR,5R,6R,6aR)-5-({[(S)-{[(S)-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}methyl)-2,6-dihydroxytetrahydrofuro[2,3-d][1,3]oxathiol-2-yl]propanoic acid, NAD-dependent lysine demalonylase and desuccinylase sirtuin-5, mitochondrial, ... | | Authors: | Zhou, Y, Hao, Q. | | Deposit date: | 2012-05-11 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Bicyclic Intermediate Structure Provides Insights into the Desuccinylation Mechanism of Human Sirtuin 5 (SIRT5)
J.Biol.Chem., 287, 2012
|
|
5VKV
 
 | |
8GUL
 
 | |
8GUM
 
 | |
6JKP
 
 | | Crystal structure of sulfoacetaldehyde reductase from Bifidobacterium kashiwanohense in complex with NAD+ | | Descriptor: | Methanol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Zhou, Y, Xu, T, Lin, L, Zhang, Y, Yuchi, Z. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.008 Å) | | Cite: | Identification and characterization of a new sulfoacetaldehyde reductase from the human gut bacteriumBifidobacterium kashiwanohense.
Biosci.Rep., 39, 2019
|
|
6JKO
 
 | | Crystal structure of sulfoacetaldehyde reductase from Bifidobacterium kashiwanohense | | Descriptor: | Methanol dehydrogenase, ZINC ION | | Authors: | Zhou, Y, Xu, T, Lin, L, Zhang, Y, Yuchi, Z. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and characterization of a new sulfoacetaldehyde reductase from the human gut bacteriumBifidobacterium kashiwanohense.
Biosci.Rep., 39, 2019
|
|
6IXJ
 
 | | The crystal structure of sulfoacetaldehyde reductase from Klebsiella oxytoca | | Descriptor: | 2-hydroxyethylsulfonic acid, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Sulfoacetaldehyde reductase | | Authors: | Zhou, Y, Xu, T, Lin, L, Zhang, Y, Yuchi, Z. | | Deposit date: | 2018-12-10 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biochemical and structural investigation of sulfoacetaldehyde reductase fromKlebsiella oxytoca.
Biochem. J., 476, 2019
|
|
5XOH
 
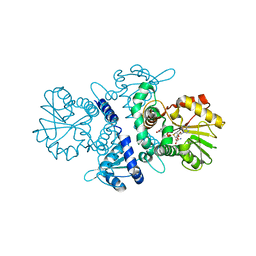 | | Crystal structure of bergaptol o-methyltransferase complex | | Descriptor: | 4-oxidanylfuro[3,2-g]chromen-7-one, Bergaptol O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zhou, Y, Zeng, Z. | | Deposit date: | 2017-05-28 | | Release date: | 2018-05-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of bergaptol o-methyltransferase complex
To Be Published
|
|
6IQL
 
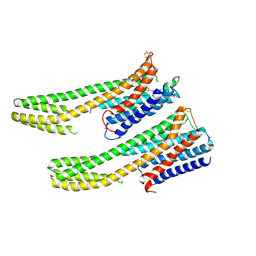 | | Crystal structure of dopamine receptor D4 bound to the subtype-selective ligand, L745870 | | Descriptor: | 3-{[4-(4-chlorophenyl)piperazin-1-yl]methyl}-1H-pyrrolo[2,3-b]pyridine, D(4) dopamine receptor,Soluble cytochrome b562,D(4) dopamine receptor | | Authors: | Zhou, Y, Cao, C, Zhang, X.C. | | Deposit date: | 2018-11-08 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of dopamine receptor D4 bound to the subtype selective ligand, L745870.
Elife, 8, 2019
|
|
6KIM
 
 | |
