3JB6
 
 | | In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, RNA-dependent RNA polymerase, VP1 CSP, ... | | Authors: | Zhang, X, Ding, K, Yu, X.K, Chang, W, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2015-08-02 | | Release date: | 2015-10-28 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus.
Nature, 527, 2015
|
|
1CDZ
 
 | | BRCT DOMAIN FROM DNA-REPAIR PROTEIN XRCC1 | | Descriptor: | PROTEIN (DNA-REPAIR PROTEIN XRCC1) | | Authors: | Zhang, X, Morera, S, Bates, P, Whitehead, P, Coffer, A, Hainbucher, K, Nash, R, Sternberg, M, Lindahl, T, Freemont, P. | | Deposit date: | 1999-03-04 | | Release date: | 2000-02-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of an XRCC1 BRCT domain: a new protein-protein interaction module.
EMBO J., 17, 1998
|
|
4JDL
 
 | |
3AH5
 
 | | Crystal Structure of flavin dependent thymidylate synthase ThyX from helicobacter pylori complexed with FAD and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Zhang, X, Zhang, J, Hu, Y, Zou, Q, Wang, D. | | Deposit date: | 2010-04-14 | | Release date: | 2011-04-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and functional analysis of a flavin dependent thymidylate synthase from helicobacter pylori
To be Published
|
|
5HWL
 
 | | Human glutathione s-transferase Mu2 complexed with BDEA, monoclinic crystal form | | Descriptor: | GLUTATHIONE, Glutathione S-transferase Mu 2, N,N'-(butane-1,4-diyl)bis{2-[2,3-dichloro-4-(2-methylidenebutanoyl)phenoxy]acetamide} | | Authors: | Zhang, X, Wei, J, Wu, S, Zhang, H.P, Luo, M, Yang, X.L, Liao, F, Wang, D.Q. | | Deposit date: | 2016-01-29 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Human glutathione s-transferase Mu2 complexed with BDEA, monoclinic crystal form
To Be Published
|
|
4JDA
 
 | | Complex structure of abscisic acid receptor PYL3 with (-)-ABA | | Descriptor: | (2Z,4E)-5-[(1R)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3 | | Authors: | Zhang, X, Wang, G, Chen, Z. | | Deposit date: | 2013-02-24 | | Release date: | 2013-07-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Insights into the Abscisic Acid Stereospecificity by the ABA Receptors PYR/PYL/RCAR
Plos One, 8, 2013
|
|
3KL1
 
 | |
3OQU
 
 | | Crystal structure of native abscisic acid receptor PYL9 with ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL9 | | Authors: | Zhang, X, Zhang, Q, Chen, Z. | | Deposit date: | 2010-09-04 | | Release date: | 2011-09-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural Insights into the Abscisic Acid Stereospecificity by the ABA Receptors PYR/PYL/RCAR
Plos One, 8, 2013
|
|
3KLX
 
 | |
3OJI
 
 | | X-ray crystal structure of the Py13 -pyrabactin complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL3, SULFATE ION | | Authors: | Zhang, X, Zhang, Q, Wang, G, Chen, Z. | | Deposit date: | 2010-08-23 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
4O68
 
 | | Structure of human cyclic GMP-AMP synthase (cGAS) | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhang, X, Chen, Z, Zhang, X.W, Chen, Z.J. | | Deposit date: | 2013-12-20 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.436 Å) | | Cite: | The Cytosolic DNA Sensor cGAS Forms an Oligomeric Complex with DNA and Undergoes Switch-like Conformational Changes in the Activation Loop.
Cell Rep, 6, 2014
|
|
4O67
 
 | | Human cyclic GMP-AMP synthase (cGAS) in complex with GAMP | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION, cGAMP | | Authors: | Zhang, X, Chen, Z, Zhang, X.W, Chen, Z.J. | | Deposit date: | 2013-12-20 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | The Cytosolic DNA Sensor cGAS Forms an Oligomeric Complex with DNA and Undergoes Switch-like Conformational Changes in the Activation Loop.
Cell Rep, 6, 2014
|
|
4O69
 
 | | Human cyclic GMP-AMP synthase (cGAS) in complex with sulfate ion | | Descriptor: | Cyclic GMP-AMP synthase, SULFATE ION, ZINC ION | | Authors: | Zhang, X, Chen, Z, Zhang, X.W, Chen, Z.J. | | Deposit date: | 2013-12-20 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | The Cytosolic DNA Sensor cGAS Forms an Oligomeric Complex with DNA and Undergoes Switch-like Conformational Changes in the Activation Loop.
Cell Rep, 6, 2014
|
|
4O6A
 
 | | Mouse cyclic GMP-AMP synthase (cGAS) in complex with DNA | | Descriptor: | Cyclic GMP-AMP synthase, DNA1, DNA2, ... | | Authors: | Zhang, X, Chen, Z, Zhang, X.W, Chen, Z.J. | | Deposit date: | 2013-12-20 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | The Cytosolic DNA Sensor cGAS Forms an Oligomeric Complex with DNA and Undergoes Switch-like Conformational Changes in the Activation Loop.
Cell Rep, 6, 2014
|
|
3IYL
 
 | | Atomic CryoEM Structure of a Nonenveloped Virus Suggests How Membrane Penetration Protein is Primed for Cell Entry | | Descriptor: | Core protein VP6, MYRISTIC ACID, Outer capsid VP4, ... | | Authors: | Zhang, X, Jin, L, Fang, Q, Hui, W, Zhou, Z.H. | | Deposit date: | 2010-02-02 | | Release date: | 2010-05-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | 3.3 A cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry.
Cell(Cambridge,Mass.), 141, 2010
|
|
3J4U
 
 | | A new topology of the HK97-like fold revealed in Bordetella bacteriophage: non-covalent chainmail secured by jellyrolls | | Descriptor: | cementing protein, major capsid protein | | Authors: | Zhang, X, Guo, H, Jin, L, Czornyj, E, Hodes, A, Hui, W.H, Nieh, A.W, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2013-10-09 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A new topology of the HK97-like fold revealed in Bordetella bacteriophage by cryoEM at 3.5 A resolution.
Elife, 2, 2013
|
|
3J9D
 
 | | Atomic structure of a non-enveloped virus reveals pH sensors for a coordinated process of cell entry | | Descriptor: | Outer capsid protein VP2, ZINC ION | | Authors: | Zhang, X, Patel, A, Celma, C, Roy, P, Zhou, Z.H. | | Deposit date: | 2015-01-09 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atomic model of a nonenveloped virus reveals pH sensors for a coordinated process of cell entry.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3J9E
 
 | | Atomic structure of a non-enveloped virus reveals pH sensors for a coordinated process of cell entry | | Descriptor: | VP5 | | Authors: | Zhang, X, Patel, A, Celma, C, Roy, P, Zhou, Z.H. | | Deposit date: | 2015-01-10 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atomic model of a nonenveloped virus reveals pH sensors for a coordinated process of cell entry.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3JB7
 
 | | In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus | | Descriptor: | CPV RNA-dependent RNA polymerase, CYTIDINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, X, Ding, K, Yu, X.K, Chang, W, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2015-08-03 | | Release date: | 2015-10-28 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (4.001 Å) | | Cite: | In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus.
Nature, 527, 2015
|
|
5GTX
 
 | | Crystal structure of mutated buckwheat glutaredoxin | | Descriptor: | buckwheat glutaredoxin | | Authors: | Zhang, X, Wang, W, Zhao, Y, Wang, Z, Wang, H. | | Deposit date: | 2016-08-23 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural insights into the binding of buckwheat glutaredoxin with GSH and regulation of its catalytic activity
J. Inorg. Biochem., 173, 2017
|
|
5YB3
 
 | | Crystal structure of HP23L/N36 | | Descriptor: | Envelope glycoprotein, HP23L | | Authors: | Zhang, X, Wang, X, He, Y. | | Deposit date: | 2017-09-03 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Structural Insights into the Mechanisms of Action of Short-Peptide HIV-1 Fusion Inhibitors Targeting the Gp41 Pocket
Front Cell Infect Microbiol, 8, 2018
|
|
5YB2
 
 | | Crystal structure of LP-11/N44 | | Descriptor: | Envelope glycoprotein, LP-11 | | Authors: | Zhang, X, Wang, X, He, Y. | | Deposit date: | 2017-09-03 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Insights into the Mechanisms of Action of Short-Peptide HIV-1 Fusion Inhibitors Targeting the Gp41 Pocket
Front Cell Infect Microbiol, 8, 2018
|
|
5YB4
 
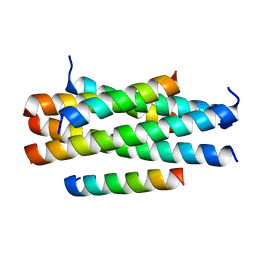 | | Crystal structure of HP23LN36KR | | Descriptor: | HP23L, N36KR | | Authors: | Zhang, X, Wang, X, He, Y. | | Deposit date: | 2017-09-03 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into the Mechanisms of Action of Short-Peptide HIV-1 Fusion Inhibitors Targeting the Gp41 Pocket
Front Cell Infect Microbiol, 8, 2018
|
|
4DSB
 
 | | Complex Structure of Abscisic Acid Receptor PYL3 with (+)-ABA in Spacegroup of I 212121 at 2.70A | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3 | | Authors: | Zhang, X, Zhang, Q, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
4DS8
 
 | | Complex structure of abscisic acid receptor PYL3-(+)-ABA-HAB1 in the presence of Mn2+ | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3, GLYCEROL, ... | | Authors: | Zhang, X, Zhang, Q, Wang, G, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
