6LG1
 
 | | Crystal structure of LpCGTa in complex with UDP | | Descriptor: | LpCGTa, URIDINE-5'-DIPHOSPHATE | | Authors: | Gao, H.M, Yun, C.H. | | Deposit date: | 2019-12-04 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.047 Å) | | Cite: | Dissection of the general two-step di- C -glycosylation pathway for the biosynthesis of (iso)schaftosides in higher plants.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6LG0
 
 | | Crystal structure of SbCGTa in complex with UDP | | Descriptor: | SbCGTa, URIDINE-5'-DIPHOSPHATE | | Authors: | Gao, H.M, Yun, C.H. | | Deposit date: | 2019-12-04 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Dissection of the general two-step di- C -glycosylation pathway for the biosynthesis of (iso)schaftosides in higher plants.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6LFN
 
 | | Crystal structure of LpCGTb | | Descriptor: | LpCGTb, PHOSPHATE ION | | Authors: | Gao, H.M, Yun, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | Dissection of the general two-step di- C -glycosylation pathway for the biosynthesis of (iso)schaftosides in higher plants.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6AIM
 
 | | Cab2 mutant H337A complex with phosphopantothenate-cysteine | | Descriptor: | N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl-L-cysteine, Phosphopantothenate--cysteine ligase CAB2 | | Authors: | Zheng, P, Zhu, Z. | | Deposit date: | 2018-08-24 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystallographic Analysis of the Catalytic Mechanism of Phosphopantothenoylcysteine Synthetase from Saccharomyces cerevisiae.
J. Mol. Biol., 431, 2019
|
|
6AI9
 
 | | Cab2 mutant-H337A complex with phosphopantothenate | | Descriptor: | N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanine, Phosphopantothenate--cysteine ligase CAB2 | | Authors: | Zheng, P, Zhu, Z. | | Deposit date: | 2018-08-22 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystallographic Analysis of the Catalytic Mechanism of Phosphopantothenoylcysteine Synthetase from Saccharomyces cerevisiae.
J. Mol. Biol., 431, 2019
|
|
6AI8
 
 | | Cab2 mutant-H337A | | Descriptor: | GLYCEROL, Phosphopantothenate--cysteine ligase CAB2, SULFATE ION | | Authors: | Zheng, P, Zhu, Z. | | Deposit date: | 2018-08-22 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic Analysis of the Catalytic Mechanism of Phosphopantothenoylcysteine Synthetase from Saccharomyces cerevisiae.
J. Mol. Biol., 431, 2019
|
|
6AIP
 
 | | Cab2 mutant-H337A complex with phosphopantothenoylcystine | | Descriptor: | (5R,12R,17R)-17-amino-12-carboxy-1,1,5-trihydroxy-4,4-dimethyl-6,10-dioxo-2-oxa-14,15-dithia-7,11-diaza-1-phosphaoctadecan-18-oic acid 1-oxide (non-preferred name), Phosphopantothenate--cysteine ligase CAB2 | | Authors: | Zheng, P, Zhu, Z. | | Deposit date: | 2018-08-24 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystallographic Analysis of the Catalytic Mechanism of Phosphopantothenoylcysteine Synthetase from Saccharomyces cerevisiae.
J. Mol. Biol., 431, 2019
|
|
6AIK
 
 | | Cab2 mutant H337A complex with phosphopantothenoyl-CMP | | Descriptor: | PHOSPHORIC ACID MONO-[3-(3-{[5-(4-AMINO-2-OXO-2H-PYRIMIDIN-1-YL)-3,4- DIHYDROXY-TETRAHYDRO-FURAN-2- YLMETHOXY]-HYDROXY-PHOSPHORYLOXY}-3-OXO-PROPYLCARBAMOYL)-3-HYDROXY-2,2- DIMETHYL-PROPYL] ESTER, Phosphopantothenate--cysteine ligase CAB2 | | Authors: | Zheng, P, Zhu, Z. | | Deposit date: | 2018-08-24 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystallographic Analysis of the Catalytic Mechanism of Phosphopantothenoylcysteine Synthetase from Saccharomyces cerevisiae.
J. Mol. Biol., 431, 2019
|
|
7CFS
 
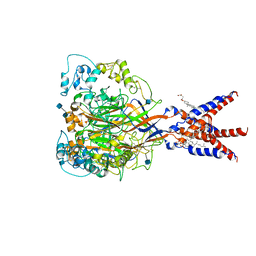 | | Cryo-EM strucutre of human acid-sensing ion channel 1a at pH 8.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Sun, D.M, Liu, S.L, Li, S.Y, Yang, F, Tian, C.L. | | Deposit date: | 2020-06-28 | | Release date: | 2020-10-21 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural insights into human acid-sensing ion channel 1a inhibition by snake toxin mambalgin1.
Elife, 9, 2020
|
|
7CFT
 
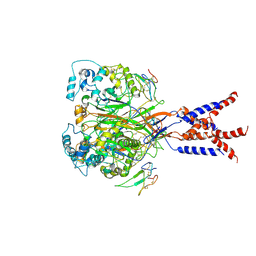 | | Cryo-EM strucutre of human acid-sensing ion channel 1a in complex with snake toxin Mambalgin1 at pH 8.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, Mambalgin-1 | | Authors: | Sun, D.M, Liu, S.L, Li, S.Y, Yang, F, Tian, C.L. | | Deposit date: | 2020-06-28 | | Release date: | 2020-10-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into human acid-sensing ion channel 1a inhibition by snake toxin mambalgin1.
Elife, 9, 2020
|
|
8GYD
 
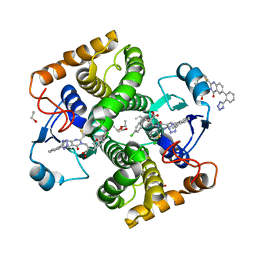 | | Structure of Schistosoma japonicum Glutathione S-transferase bound with the ligand complex of 16 | | Descriptor: | (2R)-2-[[2-(5-chloranylthiophen-2-yl)-4-oxidanylidene-6-[2-(1H-1,2,3,4-tetrazol-5-yl)phenyl]quinazolin-3-yl]methyl]-3-(4-chlorophenyl)propanoic acid, ETHANOL, Glutathione S-transferase class-mu 26 kDa isozyme | | Authors: | Wen, X, Jin, R, Hu, H, Zhu, J, Song, W, Lu, X. | | Deposit date: | 2022-09-22 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery, SAR Study of GST Inhibitors from a Novel Quinazolin-4(1 H )-one Focused DNA-Encoded Library.
J.Med.Chem., 66, 2023
|
|
5X41
 
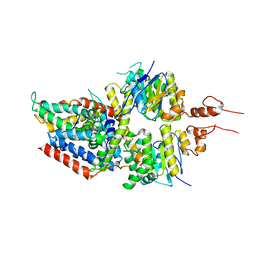 | | 3.5A resolution structure of a cobalt energy-coupling factor transporter using LCP method-CbiMQO | | Descriptor: | Cobalt ABC transporter ATP-binding protein, Cobalt transport protein CbiM, Uncharacterized protein CbiQ | | Authors: | Bao, Z, Qi, X, Zhao, W, Li, D, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
5X40
 
 | | Structure of a CbiO dimer bound with AMPPCP | | Descriptor: | Cobalt ABC transporter ATP-binding protein, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Bao, Z, Qi, X, Wang, J, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
5X3X
 
 | | 2.8A resolution structure of a cobalt energy-coupling factor transporter-CbiMQO | | Descriptor: | Cobalt ABC transporter ATP-binding protein, Cobalt transport protein CbiM, Uncharacterized protein CbiQ | | Authors: | Bao, Z, Qi, X, Wang, J, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.788 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
8HOA
 
 | | ScRIPK mutant K124R | | Descriptor: | (2R,5R)-hexane-2,5-diol, ADENOSINE-5'-TRIPHOSPHATE, FORMIC ACID, ... | | Authors: | Fang, J.L, Zhang, M.Q, Ming, Z.H. | | Deposit date: | 2022-12-09 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.676 Å) | | Cite: | Receptor-like cytoplasmic kinase ScRIPK in sugarcane regulates disease resistance and drought tolerance in Arabidopsis.
Front Plant Sci, 14, 2023
|
|
8HOD
 
 | | ScRIPK MUTANT-S253A, T254A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, Hypothetical protein | | Authors: | Fang, J.L, Zhang, M.Q. | | Deposit date: | 2022-12-09 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Receptor-like cytoplasmic kinase ScRIPK in sugarcane regulates disease resistance and drought tolerance in Arabidopsis.
Front Plant Sci, 14, 2023
|
|
8HO6
 
 | | ScRIPK WT | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ScRIPK kinase protein | | Authors: | Fang, J.L, Zhang, M.Q. | | Deposit date: | 2022-12-09 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Receptor-like cytoplasmic kinase ScRIPK in sugarcane regulates disease resistance and drought tolerance in Arabidopsis.
Front Plant Sci, 14, 2023
|
|
8IVX
 
 | | Crystal structure of NRP2 in complex with aNRP2-14 Fab fragment | | Descriptor: | 1,2-ETHANEDIOL, Heavy chain of antibody 14V4 Fab fragment, Light chain of antibody 14V4 Fab fragment, ... | | Authors: | Geng, Y, Zhai, L. | | Deposit date: | 2023-03-29 | | Release date: | 2023-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibition of VEGF binding to neuropilin-2 enhances chemosensitivity and inhibits metastasis in triple-negative breast cancer.
Sci Transl Med, 15, 2023
|
|
8IVW
 
 | | Crystal structure of NRP2 in complex with aNRP2-10 Fab fragment | | Descriptor: | Heavy chian of antibody 10V8 Fab fragment, Light chain of antibody 10V8 Fab fragment, Neuropilin-2 | | Authors: | Geng, Y, Zhai, L. | | Deposit date: | 2023-03-29 | | Release date: | 2023-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Inhibition of VEGF binding to neuropilin-2 enhances chemosensitivity and inhibits metastasis in triple-negative breast cancer.
Sci Transl Med, 15, 2023
|
|
7FIV
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidA and CidBND1-ND2 from wPip(Tunis) | | Descriptor: | CidA_I gamma/2 protein, CidB_I b/2 protein | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIT
 
 | | Crystal structure of Wolbachia cytoplasmic incompatibility factor CidA from wMel | | Descriptor: | bacteria factor 1 | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIW
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidAwMel(ST) and CidBND1-ND2 from wPip(Pel) | | Descriptor: | ULP_PROTEASE domain-containing protein, bacteria factor 4,CidA I(Zeta/1) protein | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIU
 
 | | Crystal structure of the DUB domain of Wolbachia cytoplasmic incompatibility factor CidB from wMel | | Descriptor: | ULP_PROTEASE domain-containing protein | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
6A58
 
 | | Structure of histone demethylase REF6 | | Descriptor: | Lysine-specific demethylase REF6, ZINC ION | | Authors: | Tian, Z, Chen, Z. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms.
Cell Discov, 6, 2020
|
|
5CFF
 
 | | Crystal structure of Miranda/Staufen dsRBD5 complex | | Descriptor: | Miranda, Staufen | | Authors: | Shan, Z, Wen, W. | | Deposit date: | 2015-07-08 | | Release date: | 2015-10-21 | | Last modified: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural basis of Miranda-mediated Staufen localization during Drosophila neuroblast asymmetric division
Nat Commun, 6, 2015
|
|
