4E7G
 
 | |
4E7F
 
 | | E. cloacae C115D MurA in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, ... | | Authors: | Zhu, J.-Y, Betzi, S, Yang, Y, Schonbrunn, E. | | Deposit date: | 2012-03-16 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Open-close transition of MurA
To be Published
|
|
4E7B
 
 | |
4E7E
 
 | |
1Y7H
 
 | | Structural and biochemical studies identify tobacco SABP2 as a methylsalicylate esterase and further implicate it in plant innate immunity, Northeast Structural Genomics Target AR2241 | | Descriptor: | THIOCYANATE ION, salicylic acid-binding protein 2 | | Authors: | Forouhar, F, Yang, Y, Kumar, D, Chen, Y, Fridman, E, Park, S.W, Chiang, Y, Acton, T.B, Montelione, G.T, Pichersky, E, Klessig, D.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-12-08 | | Release date: | 2004-12-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1Y7I
 
 | | Structural and biochemical studies identify tobacco SABP2 as a methylsalicylate esterase and further implicate it in plant innate immunity, Northeast Structural Genomics Target AR2241 | | Descriptor: | 2-HYDROXYBENZOIC ACID, salicylic acid-binding protein 2 | | Authors: | Forouhar, F, Yang, Y, Kumar, D, Chen, Y, Fridman, E, Park, S.W, Chiang, Y, Acton, T.B, Montelione, G.T, Pichersky, E, Klessig, D.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-12-08 | | Release date: | 2004-12-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
3TCL
 
 | | Crystal Structure of HIV-1 Neutralizing Antibody CH04 | | Descriptor: | CH04 Heavy Chain Fab, CH04 Light Chain Fab, IMIDAZOLE | | Authors: | Louder, R.K, McLellan, J.S, Pancera, M, Yang, Y, Zhang, B, Bonsignori, M, Kwong, P.D. | | Deposit date: | 2011-08-09 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
7EW9
 
 | | GDP-bound KRAS G12D in complex with TH-Z816 | | Descriptor: | 7-(8-methylnaphthalen-1-yl)-4-[(2~{R})-2-methylpiperazin-1-yl]-2-[[(2~{S})-1-methylpyrrolidin-2-yl]methoxy]-6,8-dihydro-5~{H}-pyrido[3,4-d]pyrimidine, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Shen, P, Yang, Y, Yang, Y, Zhang, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | KRAS(G12D) can be targeted by potent inhibitors via formation of salt bridge.
Cell Discov, 8, 2022
|
|
7EWB
 
 | | GDP-bound KRAS G12D in complex with TH-Z835 | | Descriptor: | 4-[(1~{S},5~{R})-3,8-diazabicyclo[3.2.1]octan-3-yl]-7-(8-methylnaphthalen-1-yl)-2-[[(2~{S})-1-methylpyrrolidin-2-yl]methoxy]-6,8-dihydro-5~{H}-pyrido[3,4-d]pyrimidine, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Shen, P, Yang, Y, Yang, Y, Zhang, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | KRAS(G12D) can be targeted by potent inhibitors via formation of salt bridge.
Cell Discov, 8, 2022
|
|
7EWA
 
 | | GDP-bound KRAS G12D in complex with TH-Z827 | | Descriptor: | 4-[(1~{R},5~{S})-3,8-diazabicyclo[3.2.1]octan-8-yl]-7-(8-methylnaphthalen-1-yl)-2-[[(2~{S})-1-methylpyrrolidin-2-yl]methoxy]-6,8-dihydro-5~{H}-pyrido[3,4-d]pyrimidine, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Shen, P, Yang, Y, Yang, Y, Zhang, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | KRAS(G12D) can be targeted by potent inhibitors via formation of salt bridge.
Cell Discov, 8, 2022
|
|
3BU8
 
 | | Crystal Structure of TRF2 TRFH domain and TIN2 peptide complex | | Descriptor: | TERF1-interacting nuclear factor 2, Telomeric repeat-binding factor 2 | | Authors: | Chen, Y, Yang, Y, van Overbeek, M, Donigian, J.R, Baciu, P, de Lange, T, Lei, M. | | Deposit date: | 2008-01-02 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins.
Science, 319, 2008
|
|
7QUR
 
 | | SARS-CoV-2 Spike with ethylbenzamide-tri-iodo Siallyllactose, C3 symmetry | | Descriptor: | 2,3,5-tris(iodanyl)benzamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Naismith, J.H, Yang, Y, Liu, J.W. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-01 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Pathogen-sugar interactions revealed by universal saturation transfer analysis.
Science, 377, 2022
|
|
7QUS
 
 | | SARS-CoV-2 Spike, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Naismith, J.H, Yang, Y, Liu, J.W. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-08 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Pathogen-sugar interactions revealed by universal saturation transfer analysis.
Science, 377, 2022
|
|
3BUA
 
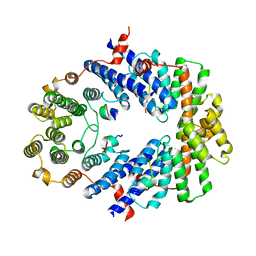 | | Crystal Structure of TRF2 TRFH domain and APOLLO peptide complex | | Descriptor: | DNA cross-link repair 1B protein, Telomeric repeat-binding factor 2 | | Authors: | Chen, Y, Yang, Y, van Overbeek, M, Donigian, J.R, Baciu, P, de Lange, T, Lei, M. | | Deposit date: | 2008-01-02 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins.
Science, 319, 2008
|
|
3BQO
 
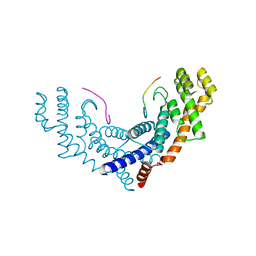 | | Crystal Structure of TRF1 TRFH domain and TIN2 peptide complex | | Descriptor: | TERF1-interacting nuclear factor 2, Telomeric repeat-binding factor 1 | | Authors: | Chen, Y, Yang, Y, van Overbeek, M, Donigian, J.R, Baciu, P, de Lange, T, Lei, M. | | Deposit date: | 2007-12-20 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins.
Science, 319, 2008
|
|
1M6E
 
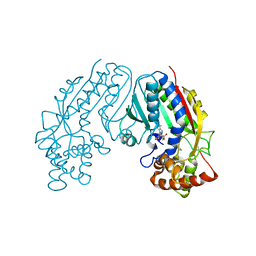 | | CRYSTAL STRUCTURE OF SALICYLIC ACID CARBOXYL METHYLTRANSFERASE (SAMT) | | Descriptor: | 2-HYDROXYBENZOIC ACID, LUTETIUM (III) ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Zubieta, C, Ross, J.R, Koscheski, P, Yang, Y, Pichersky, E, Noel, J.P. | | Deposit date: | 2002-07-16 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Substrate Recognition in The Salicylic Acid Carboxyl Methyltransferase Family
Plant Cell, 15, 2003
|
|
4GKG
 
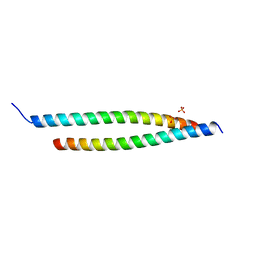 | | Crystal structure of the S-Helix Linker | | Descriptor: | C4-dicarboxylate transport sensor protein dctB, PHOSPHATE ION | | Authors: | Liu, J.W, Lu, D, Sun, Y.J, Wen, J, Yang, Y, Yang, J.G, Wei, X.L, Zhang, X.D, Wang, Y.P. | | Deposit date: | 2012-08-11 | | Release date: | 2013-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Crystal structure of the S-Helix Linker
To be Published
|
|
2RFS
 
 | | X-ray structure of SU11274 bound to c-Met | | Descriptor: | Hepatocyte growth factor receptor, N-(3-chlorophenyl)-N-methyl-2-oxo-3-[(3,4,5-trimethyl-1H-pyrrol-2-yl)methyl]-2H-indole-5-sulfonamide | | Authors: | Bellon, S.F, Kaplan-Lefko, P, Yang, Y, Zhang, Y, Moriguchi, J, Dussault, I. | | Deposit date: | 2007-10-01 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | c-Met inhibitors with novel binding mode show activity against several hereditary papillary renal cell carcinoma-related mutations.
J.Biol.Chem., 283, 2008
|
|
2RFN
 
 | | x-ray structure of c-Met with inhibitor. | | Descriptor: | 2-benzyl-5-(3-fluoro-4-{[6-methoxy-7-(3-morpholin-4-ylpropoxy)quinolin-4-yl]oxy}phenyl)-3-methylpyrimidin-4(3H)-one, Hepatocyte growth factor receptor | | Authors: | Bellon, S.F, Kaplan-Lefko, P, Yang, Y, Zhang, Y, Moriguchi, J, Dussault, I. | | Deposit date: | 2007-10-01 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | c-Met inhibitors with novel binding mode show activity against several hereditary papillary renal cell carcinoma-related mutations.
J.Biol.Chem., 283, 2008
|
|
1NI8
 
 | | H-NS dimerization motif | | Descriptor: | DNA-binding protein H-NS | | Authors: | Bloch, V, Yang, Y, Margeat, E, Chavanieu, A, Aug, M.T, Robert, B, Arold, S, Rimsky, S, Kochoyan, M. | | Deposit date: | 2002-12-22 | | Release date: | 2003-02-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The H-NS dimerisation domain defines a new fold contributing to DNA recognition
Nat.Struct.Biol., 10, 2003
|
|
5DAY
 
 | | The structure of NAP1-Related Protein(NRP1) in Arabidopsis | | Descriptor: | CALCIUM ION, NAP1-related protein 1 | | Authors: | Zhu, Y, Rong, L, Yang, Y, Zhang, C, Feng, H.Y, Zheng, L.N, Shen, W.H, Ma, J.B, Dong, A.W. | | Deposit date: | 2015-08-20 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.329 Å) | | Cite: | The structure of NAP1-Related Protein(NRP1) in Arabidopsis
To Be Published
|
|
6PQY
 
 | | Cryo-EM structure of HzTransib/TIR DNA transposon end complex (TEC) | | Descriptor: | DNA (5'-D(P*CP*AP*CP*GP*GP*TP*GP*GP*AP*TP*CP*GP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*CP*GP*AP*TP*CP*CP*AP*CP*CP*GP*TP*G)-3'), Putative DNA-mediated transposase | | Authors: | Liu, C, Yang, Y, Schatz, D.G. | | Deposit date: | 2019-07-10 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structures of a RAG-like transposase during cut-and-paste transposition.
Nature, 575, 2019
|
|
6PR5
 
 | | Cryo-EM structure of HzTransib strand transfer complex (STC) | | Descriptor: | DNA (30-MER), DNA (39-MER), DNA (5'-D(*GP*AP*TP*CP*TP*GP*GP*CP*CP*TP*AP*GP*AP*TP*CP*TP*CP*A)-3'), ... | | Authors: | Liu, C, Yang, Y, Schatz, D.G. | | Deposit date: | 2019-07-10 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of a RAG-like transposase during cut-and-paste transposition.
Nature, 575, 2019
|
|
6PQN
 
 | | Crystal structure of HzTransib transposase | | Descriptor: | GLYCEROL, PHOSPHATE ION, Putative DNA-mediated transposase, ... | | Authors: | Liu, C, Yang, Y, Schatz, D.G. | | Deposit date: | 2019-07-09 | | Release date: | 2019-10-09 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structures of a RAG-like transposase during cut-and-paste transposition.
Nature, 575, 2019
|
|
6PQU
 
 | | Cryo-EM structure of HzTransib/nicked TIR substrate DNA pre-reaction complex (PRC) | | Descriptor: | DNA (5'-D(P*AP*TP*CP*TP*GP*GP*CP*CP*TP*AP*GP*AP*TP*CP*T)-3'), DNA (5'-D(P*CP*AP*CP*GP*GP*TP*GP*GP*AP*TP*CP*GP*AP*AP*AP*A)-3'), DNA-mediated transposase, ... | | Authors: | Liu, C, Yang, Y, Schatz, D.G. | | Deposit date: | 2019-07-10 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of a RAG-like transposase during cut-and-paste transposition.
Nature, 575, 2019
|
|
