7Y0L
 
 | | SulE-S209A | | 分子名称: | Alpha/beta fold hydrolase, GLYCEROL, METHYL 2-[({[(4-METHOXY-6-METHYL-1,3,5-TRIAZIN-2-YL)AMINO]CARBONYL}AMINO)SULFONYL]BENZOATE | | 著者 | Liu, B, Ran, T, He, J, Wang, W. | | 登録日 | 2022-06-05 | | 公開日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8HQ3
 
 | | KL1 in complex with CRM1-Ran-RanBP1 | | 分子名称: | CHLORIDE ION, CRM1 isoform 1, DIMETHYL SULFOXIDE, ... | | 著者 | Sun, Q, Jian, L. | | 登録日 | 2022-12-13 | | 公開日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Discovery of Aminoratjadone Derivatives as Potent Noncovalent CRM1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
8HQ6
 
 | | KL2 in complex with CRM1-Ran-RanBP1 | | 分子名称: | CHLORIDE ION, CRM1 isoform 1, DIMETHYL SULFOXIDE, ... | | 著者 | Sun, Q, Jian, L. | | 登録日 | 2022-12-13 | | 公開日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Discovery of Aminoratjadone Derivatives as Potent Noncovalent CRM1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
8J4T
 
 | | GajA-GajB complex | | 分子名称: | Endonuclease GajA, Gabija protein GajB | | 著者 | Wei, T, Ma, J, Huo, Y. | | 登録日 | 2023-04-20 | | 公開日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural and biochemical insights into the mechanism of the Gabija bacterial immunity system.
Nat Commun, 15, 2024
|
|
5WKT
 
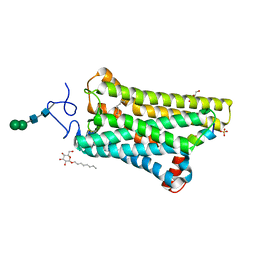 | | 3.2-Angstrom In situ Mylar structure of bovine opsin at 100 K | | 分子名称: | Rhodopsin, SULFATE ION, Transducin Galpha peptide, ... | | 著者 | Broecker, J, Morizumi, T, Ou, W.-L, Ernst, O.P. | | 登録日 | 2017-07-25 | | 公開日 | 2017-12-13 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | High-throughput in situ X-ray screening of and data collection from protein crystals at room temperature and under cryogenic conditions.
Nat Protoc, 13, 2018
|
|
5XBW
 
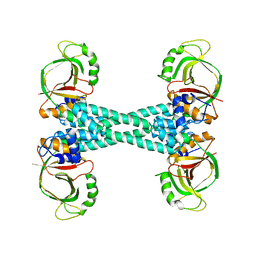 | | The structure of BrlR | | 分子名称: | Probable transcriptional regulator | | 著者 | Wang, F, Qing, H, Gu, L. | | 登録日 | 2017-03-21 | | 公開日 | 2018-05-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.109 Å) | | 主引用文献 | BrlR from Pseudomonas aeruginosa is a receptor for both cyclic di-GMP and pyocyanin.
Nat Commun, 9, 2018
|
|
5XBI
 
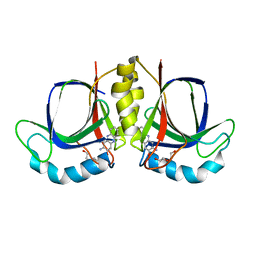 | |
5XBT
 
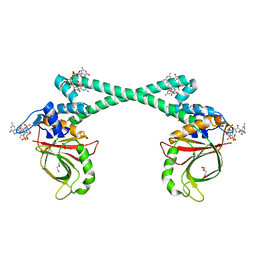 | | The structure of BrlR bound to c-di-GMP | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Wang, F, Qing, H, Gu, L. | | 登録日 | 2017-03-21 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.495 Å) | | 主引用文献 | BrlR from Pseudomonas aeruginosa is a receptor for both cyclic di-GMP and pyocyanin.
Nat Commun, 9, 2018
|
|
5XSN
 
 | | The catalytic domain of GdpP with c-di-AMP | | 分子名称: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MANGANESE (II) ION, Phosphodiesterase acting on cyclic dinucleotides | | 著者 | Wang, F, Gu, L. | | 登録日 | 2017-06-14 | | 公開日 | 2018-01-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.501 Å) | | 主引用文献 | Structural and biochemical characterization of the catalytic domains of GdpP reveals a unified hydrolysis mechanism for the DHH/DHHA1 phosphodiesterase
Biochem. J., 475, 2018
|
|
5XSI
 
 | | The catalytic domain of GdpP | | 分子名称: | MANGANESE (II) ION, Phosphodiesterase acting on cyclic dinucleotides | | 著者 | Wang, F, Gu, L. | | 登録日 | 2017-06-14 | | 公開日 | 2018-01-31 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural and biochemical characterization of the catalytic domains of GdpP reveals a unified hydrolysis mechanism for the DHH/DHHA1 phosphodiesterase
Biochem. J., 475, 2018
|
|
5XSP
 
 | | The catalytic domain of GdpP with 5'-pApA | | 分子名称: | ADENOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, Phosphodiesterase acting on cyclic dinucleotides | | 著者 | Wang, F, Gu, L. | | 登録日 | 2017-06-15 | | 公開日 | 2018-01-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.146 Å) | | 主引用文献 | Structural and biochemical characterization of the catalytic domains of GdpP reveals a unified hydrolysis mechanism for the DHH/DHHA1 phosphodiesterase
Biochem. J., 475, 2018
|
|
5XT3
 
 | | The catalytic domain of GdpP with c-di-GMP | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MANGANESE (II) ION, Phosphodiesterase acting on cyclic dinucleotides | | 著者 | Wang, F, Gu, L. | | 登録日 | 2017-06-16 | | 公開日 | 2018-01-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.591 Å) | | 主引用文献 | Structural and biochemical characterization of the catalytic domains of GdpP reveals a unified hydrolysis mechanism for the DHH/DHHA1 phosphodiesterase
Biochem. J., 475, 2018
|
|
6A06
 
 | | Structure of pSTING complex | | 分子名称: | SULFATE ION, Stimulator of interferon genes protein, cGAMP | | 著者 | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | 登録日 | 2018-06-05 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.792 Å) | | 主引用文献 | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
6A04
 
 | | Structure of pSTING complex | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), SULFATE ION, Stimulator of interferon genes protein | | 著者 | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | 登録日 | 2018-06-05 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
6A05
 
 | | Structure of pSTING complex | | 分子名称: | 2-amino-9-[(2R,3R,3aR,5S,7aS,9R,10R,10aR,12R,14aS)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, SULFATE ION, Stimulator of interferon genes protein | | 著者 | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | 登録日 | 2018-06-05 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
6A03
 
 | | Structure of pSTING complex | | 分子名称: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, SULFATE ION, Stimulator of interferon genes protein | | 著者 | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | 登録日 | 2018-06-05 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.597 Å) | | 主引用文献 | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
7DO4
 
 | | Crystal structure of CD97-CD55 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Niu, M, Song, G. | | 登録日 | 2020-12-11 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis for CD97 recognition of the decay-accelerating factor CD55 suggests mechanosensitive activation of adhesion GPCRs.
J.Biol.Chem., 296, 2021
|
|
7EAK
 
 | | Echovirus3 full particle in complex with 5G3 fab | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | 著者 | Feng, R. | | 登録日 | 2021-03-07 | | 公開日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural basis for neutralization of an anicteric hepatitis associated echovirus by a potent neutralizing antibody.
Cell Discov, 7, 2021
|
|
7EAH
 
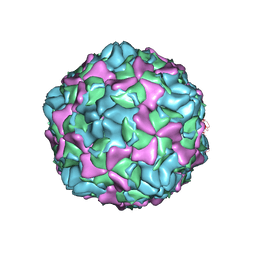 | | Echovirus3 empty expanded particle | | 分子名称: | Capsid protein VP0, Capsid protein VP1, Capsid protein VP3 | | 著者 | Feng, R. | | 登録日 | 2021-03-07 | | 公開日 | 2021-07-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis for neutralization of an anicteric hepatitis associated echovirus by a potent neutralizing antibody.
Cell Discov, 7, 2021
|
|
7EAI
 
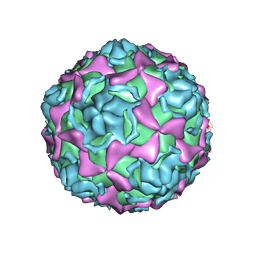 | | Echovirus3 empty compacted particle | | 分子名称: | Capsid protein VP0, Capsid protein VP1, Capsid protein VP3, ... | | 著者 | Feng, R. | | 登録日 | 2021-03-07 | | 公開日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis for neutralization of an anicteric hepatitis associated echovirus by a potent neutralizing antibody.
Cell Discov, 7, 2021
|
|
7EAJ
 
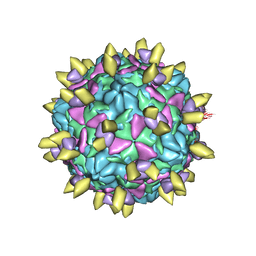 | |
7JQ2
 
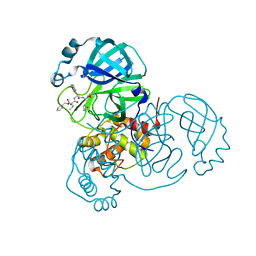 | |
7JNT
 
 | |
7JPY
 
 | |
7JQ4
 
 | |
