2AMN
 
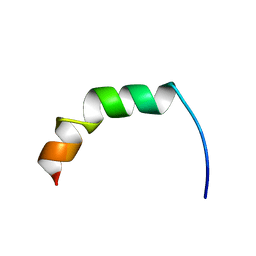 | | Solution structure of Fowlicidin-1, a novel Cathelicidin antimicrobial peptide from chicken | | Descriptor: | cathelicidin | | Authors: | Xiao, Y, Dai, H, Bommineni, Y.R, Prakash, O, Zhang, G. | | Deposit date: | 2005-08-09 | | Release date: | 2006-07-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure-activity relationships of fowlicidin-1, a cathelicidin antimicrobial peptide in chicken.
Febs J., 273, 2006
|
|
6C66
 
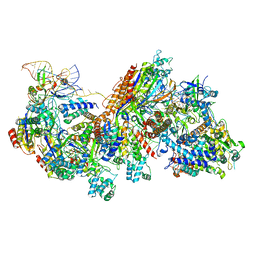 | | CRISPR RNA-guided surveillance complex, pre-nicking | | Descriptor: | CRISPR-associated helicase, Cas3 family, CRISPR-associated protein, ... | | Authors: | Xiao, Y, Luo, M, Liao, M, Ke, A. | | Deposit date: | 2018-01-17 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structure basis for RNA-guided DNA degradation by Cascade and Cas3.
Science, 361, 2018
|
|
3UB0
 
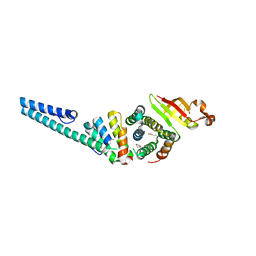 | | Crystal structure of the nonstructural protein 7 and 8 complex of Feline Coronavirus | | Descriptor: | Non-structural protein 6, nsp6,, Non-structural protein 7, ... | | Authors: | Xiao, Y, Hilgenfeld, R, Ma, Q. | | Deposit date: | 2011-10-22 | | Release date: | 2012-02-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nonstructural proteins 7 and 8 of feline coronavirus form a 2:1 heterotrimer that exhibits primer-independent RNA polymerase activity.
J.Virol., 86, 2012
|
|
2MXU
 
 | | 42-Residue Beta Amyloid Fibril | | Descriptor: | Amyloid beta A4 protein | | Authors: | Xiao, Y, Ma, B, McElheny, D, Parthasarathy, S, Long, F, Hoshi, M, Nussinov, R, Ishii, Y. | | Deposit date: | 2015-01-14 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | A beta (1-42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer's disease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3CSZ
 
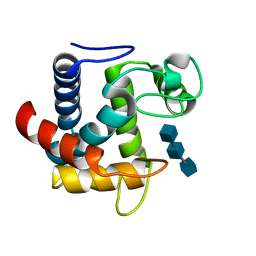 | |
3CT5
 
 | |
3CSQ
 
 | |
3CT0
 
 | |
3CSR
 
 | |
3CT1
 
 | |
3OY7
 
 | |
3OY2
 
 | |
3GQA
 
 | |
3GQK
 
 | |
3GQ9
 
 | |
3GQ7
 
 | |
3GQ8
 
 | |
3GQH
 
 | |
5XVN
 
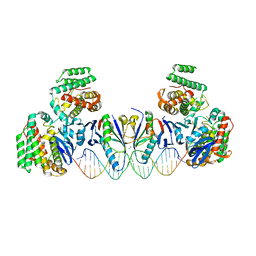 | | E. far Cas1-Cas2/prespacer binary complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (28-MER), ... | | Authors: | Xiao, Y, Ng, S, Nam, K.H, Ke, A. | | Deposit date: | 2017-06-28 | | Release date: | 2017-10-04 | | Last modified: | 2022-10-12 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | How type II CRISPR-Cas establish immunity through Cas1-Cas2-mediated spacer integration.
Nature, 550, 2017
|
|
5XVP
 
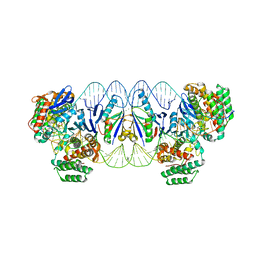 | | E. fae Cas1-Cas2/prespacer/target ternary complex revealing the fully integrated states | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (5'-D(P*TP*TP*CP*TP*CP*CP*GP*AP*G)-3'), ... | | Authors: | Xiao, Y, Ng, S, Nam, K.H, Ke, A. | | Deposit date: | 2017-06-28 | | Release date: | 2017-10-04 | | Last modified: | 2022-10-12 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | How type II CRISPR-Cas establish immunity through Cas1-Cas2-mediated spacer integration.
Nature, 550, 2017
|
|
5XVO
 
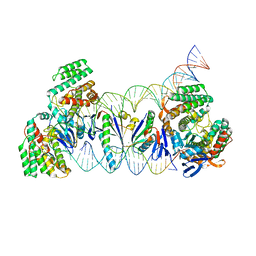 | | E. fae Cas1-Cas2/prespacer/target ternary complex revealing DNA sampling and half-integration states | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (28-MER), ... | | Authors: | Xiao, Y, Ng, S, Nam, K.H, Ke, A. | | Deposit date: | 2017-06-28 | | Release date: | 2017-10-04 | | Last modified: | 2022-10-12 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How type II CRISPR-Cas establish immunity through Cas1-Cas2-mediated spacer integration.
Nature, 550, 2017
|
|
3R04
 
 | | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors | | Descriptor: | 5-{6-[(trans-4-aminocyclohexyl)amino]pyrazin-2-yl}-1-benzofuran-2-carboxylic acid, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase pim-1 | | Authors: | Xiang, Y, Hirth, B, Asmussen, G, Biemann, H.-P, Good, A, Fitzgerald, M, Gladysheva, T, Jancsics, K, Liu, J, Metz, M, Papoulis, A, Skerlj, R, Stepp, D.J, Wei, R.R. | | Deposit date: | 2011-03-07 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3R02
 
 | | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors | | Descriptor: | 7-[(cis-4-aminocyclohexyl)amino]-5-bromo-1-benzofuran-2-carboxylic acid, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase pim-1 | | Authors: | Xiang, Y, Hirth, B, Asmussen, G, Biemann, H.-P, Good, A, Fitzgerald, M, Gladysheva, T, Jancsics, K, Liu, J, Metz, M, Papoulis, A, Skerlj, R, Stepp, D.J, Wei, R.R. | | Deposit date: | 2011-03-07 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3QC7
 
 | |
3SUC
 
 | |
