2F8P
 
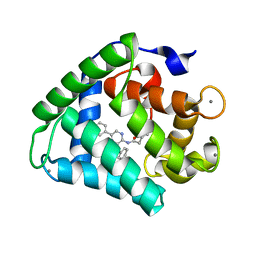 | | Crystal structure of obelin following Ca2+ triggered bioluminescence suggests neutral coelenteramide as the primary excited state | | Descriptor: | CALCIUM ION, N-[3-BENZYL-5-(4-HYDROXYPHENYL)PYRAZIN-2-YL]-2-(4-HYDROXYPHENYL)ACETAMIDE, Obelin | | Authors: | Liu, Z.J, Stepanyuk, G.A, Vysotski, E.S, Lee, J, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-12-03 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of obelin after Ca2+-triggered bioluminescence suggests neutral coelenteramide as the primary excited state.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4WBX
 
 | | Conserved hypothetical protein PF1771 from Pyrococcus furiosus solved by sulfur SAD using Swiss Light Source data | | Descriptor: | 2-keto acid:ferredoxin oxidoreductase subunit alpha | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-09-04 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
1LKP
 
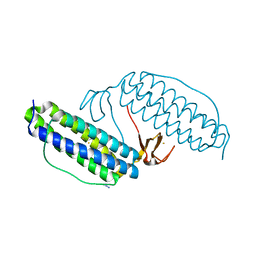 | | Crystal structure of Desulfovibrio vulgaris rubrerythrin all-iron(II) form, azide adduct | | Descriptor: | AZIDE ION, FE (II) ION, Rubrerythrin | | Authors: | Jin, S, Kurtz Jr, D.M, Liu, Z.J, Rose, J, Wang, B.C. | | Deposit date: | 2002-04-25 | | Release date: | 2002-09-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | X-ray Crystal Structures of Reduced Rubrerythrin and its Azide Adduct: A Structure-Based Mechanism for a Non-Heme Diiron Peroxidase
J.Am.Chem.Soc., 124, 2002
|
|
2QUF
 
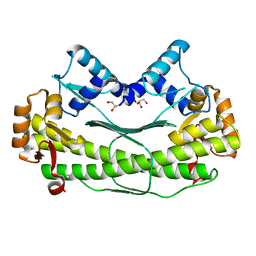 | | Crystal Structure of Transcription Factor AXXA-PF0095 from Pyrococcus furiosus | | Descriptor: | GLYCEROL, Transcription Factor PF0095 | | Authors: | Yang, H, Lipscomb, G.L, Scott, R.A, Rose, J.P, Wang, B.C. | | Deposit date: | 2007-08-05 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Transcription Factor AXXA-PF0095 from Pyrococcus furiosus
To be Published
|
|
2BN2
 
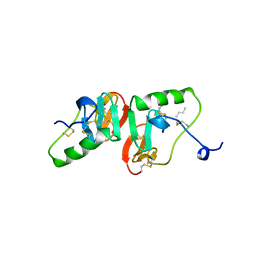 | |
2RHE
 
 | |
1AD3
 
 | |
3R7C
 
 | | The structure of a hexahestidine-tagged form of augmenter of liver regeneration reveals a novel Cd(2)Cl(4)O(6) cluster that aids in crystal packing | | Descriptor: | CADMIUM ION, CHLORIDE ION, FAD-linked sulfhydryl oxidase ALR, ... | | Authors: | Florence, Q.J, Wu, C.-K, Swindell II, J.T, Wang, B.C, Rose, J.P. | | Deposit date: | 2011-03-22 | | Release date: | 2012-03-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of a hexahestidine-tagged form of augmenter of liver regeneration reveals a novel Cd(2)Cl(4)O(6) cluster that aids in crystal packing
To be Published
|
|
2I0X
 
 | | Hypothetical protein PF1117 from Pyrococcus furiosus | | Descriptor: | Hypothetical protein PF1117 | | Authors: | Chen, L.Q, Fu, Z.-Q, Liu, Z.-J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-08-11 | | Release date: | 2006-10-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Hypothetical Protein Pf1117 from Pyrococcus furiosus
To be Published
|
|
1VK1
 
 | | Conserved hypothetical protein from Pyrococcus furiosus Pfu-392566-001 | | Descriptor: | Conserved hypothetical protein, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Shah, A, Liu, Z.J, Tempel, W, Chen, L, Lee, D, Yang, H, Chang, J, Zhao, M, Ng, J, Rose, J, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-04-13 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | (NZ)CH...O contacts assist crystallization of a ParB-like nuclease.
Bmc Struct.Biol., 7, 2007
|
|
6EB0
 
 | | STRUCTURE OF 4-HYDROXYPHENYLACETATE 3-MONOOXYGENASE (HPAB), OXYGENASE COMPONENT FROM ESCHERICHIA COLI | | Descriptor: | 4-hydroxyphenylacetate 3-monooxygenase, oxygenase subunit, ACETATE ION | | Authors: | Zhou, D, Kandavelu, P, Zhang, H, Wang, B.C, Yan, Y, Rose, J. | | Deposit date: | 2018-08-03 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural Insights into Catalytic Versatility of the Flavin-dependent Hydroxylase (HpaB) from Escherichia coli.
Sci Rep, 9, 2019
|
|
1SL7
 
 | | Crystal structure of calcium-loaded apo-obelin from Obelia longissima | | Descriptor: | CALCIUM ION, Obelin | | Authors: | Deng, L, Markova, S.V, Vysotski, E.S, Liu, Z.J, Lee, J, Rose, J, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-05 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | All three Ca2+-binding loops of photoproteins bind calcium ions: The crystal structures of calcium-loaded apo-aequorin and apo-obelin.
Protein Sci., 14, 2005
|
|
1SL8
 
 | | Calcium-loaded apo-aequorin from Aequorea victoria | | Descriptor: | Aequorin 1, CALCIUM ION | | Authors: | Deng, L, Markova, S.V, Vysotski, E.S, Liu, Z.J, Lee, J, Rose, J, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-05 | | Release date: | 2004-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | All three Ca2+-binding loops of photoproteins bind calcium ions: The crystal structures of calcium-loaded apo-aequorin and apo-obelin.
Protein Sci., 14, 2005
|
|
1EL4
 
 | | STRUCTURE OF THE CALCIUM-REGULATED PHOTOPROTEIN OBELIN DETERMINED BY SULFUR SAS | | Descriptor: | C2-HYDROXY-COELENTERAZINE, CHLORIDE ION, OBELIN | | Authors: | Liu, Z.J, Vysotski, E.S, Rose, J, Lee, J, Wang, B.C. | | Deposit date: | 2000-03-13 | | Release date: | 2001-03-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure of the Ca2+-regulated photoprotein obelin at 1.7 A resolution determined directly from its sulfur substructure.
Protein Sci., 9, 2000
|
|
1WWI
 
 | | Crystal structure of ttk003001566 from Thermus Thermophilus HB8 | | Descriptor: | hypothetical protein TTHA1479 | | Authors: | Wang, H, Murayama, K, Terada, T, Chen, L, Liu, Z.J, Wang, B.C, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-05 | | Release date: | 2005-07-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of ttk003001566 from Thermus Thermophilus HB8
TO BE PUBLISHED
|
|
2CSL
 
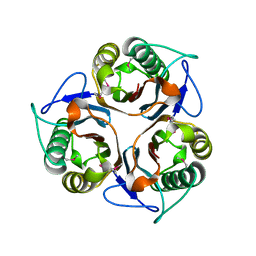 | | Crystal structure of TTHA0137 from Thermus Thermophilus HB8 | | Descriptor: | protein translation initiation inhibitor | | Authors: | Wang, H, Murayama, K, Terada, T, Chen, L, Jin, Z, Chrzas, J, Liu, Z.J, Wang, B.C, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-22 | | Release date: | 2005-11-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of TTHA0137 from Thermus Thermophilus HB8
To be Published
|
|
2DY1
 
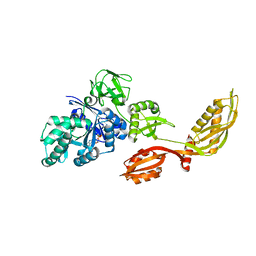 | | Crystal structure of EF-G-2 from Thermus thermophilus | | Descriptor: | Elongation factor G, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Wang, H, Takemoto, C, Murayama, K, Terada, T, Chen, L, Liu, Z.J, Wang, B.C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-04 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of EF-G-2 from Thermus thermophilus
To be Published
|
|
1QV0
 
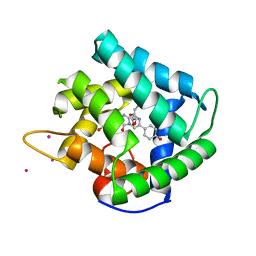 | | Atomic resolution structure of obelin from Obelia longissima | | Descriptor: | C2-HYDROPEROXY-COELENTERAZINE, COBALT (II) ION, GLYCEROL, ... | | Authors: | Liu, Z.J, Vysotski, E.S, Deng, L, Lee, J, Rose, J, Wang, B.C. | | Deposit date: | 2003-08-26 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution structure of obelin: soaking with calcium enhances electron density of the second oxygen atom substituted at the C2-position of coelenterazine.
Biochem.Biophys.Res.Commun., 311, 2003
|
|
1R7J
 
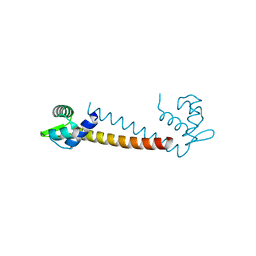 | | Crystal structure of the DNA-binding protein Sso10a from Sulfolobus solfataricus | | Descriptor: | Conserved hypothetical protein Sso10a | | Authors: | Chen, L, Chen, L.R, Zhou, X.E, Wang, Y, Kahsai, M.A, Clark, A.T, Edmondson, S.P, Liu, Z.-J, Rose, J.P, Wang, B.C, Shriver, J.W, Meehan, E.J, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-10-21 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The hyperthermophile protein Sso10a is a dimer of winged helix DNA-binding domains linked by an antiparallel coiled coil rod.
J.Mol.Biol., 341, 2004
|
|
1QV1
 
 | | Atomic resolution structure of obelin from Obelia longissima | | Descriptor: | C2-HYDROPEROXY-COELENTERAZINE, CALCIUM ION, COBALT (II) ION, ... | | Authors: | Liu, Z.J, Vysotski, E.S, Deng, L, Lee, J, Rose, J, Wang, B.C. | | Deposit date: | 2003-08-26 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution structure of obelin: soaking with calcium enhances electron density of the second oxygen atom substituted at the C2-position of coelenterazine.
Biochem.Biophys.Res.Commun., 311, 2003
|
|
3O3K
 
 | | Crystal structure of AF1382 from Archaeoglobus fulgidus | | Descriptor: | Uncharacterized protein AF_1382 | | Authors: | Zhu, J.-Y, Zhao, M, Fu, Z.-Q, Yang, H, Chang, J, Xu, H, Chen, L, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2010-07-24 | | Release date: | 2010-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Archaeoglobus fulgidus orphan ORF AF1382 determined by sulfur SAD from a moderately diffracting crystal.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3OV8
 
 | | Crystal structure of AF1382 from Archaeoglobus fulgidus, High resolution | | Descriptor: | ACETATE ION, CHLORIDE ION, Protein AF_1382 | | Authors: | Zhu, J.-Y, Zhao, M, Fu, Z.-Q, Yang, H, Chang, J, Hao, X, Chen, L, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2010-09-16 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8501 Å) | | Cite: | Structure of the Archaeoglobus fulgidus orphan ORF AF1382 determined by sulfur SAD from a moderately diffracting crystal.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2QLZ
 
 | |
2QVO
 
 | | Crystal structure of AF1382 from Archaeoglobus fulgidus | | Descriptor: | Uncharacterized protein AF_1382 | | Authors: | Zhu, J, Zhao, M, Fu, Z.-Q, Yang, H, Chang, J, Hao, X, Chen, L, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2007-08-08 | | Release date: | 2007-09-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of AF1382 from Archaeoglobus fulgidus.
To be Published
|
|
4RNP
 
 | | BACTERIOPHAGE T7 RNA POLYMERASE, HIGH SALT CRYSTAL FORM, LOW TEMPERATURE DATA, ALPHA-CARBONS ONLY | | Descriptor: | RNA POLYMERASE | | Authors: | Liu, Z.J, Sousa, R, Rose, J.P, Wang, B.C. | | Deposit date: | 1997-09-11 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of bacteriophage T7 RNA polymerase at 3.3 A resolution.
Nature, 364, 1993
|
|
