8C0I
 
 | | Structure of E. coli Class 2 L-asparaginase EcAIII, mutant M200L (acyl-enzyme intermediate) | | Descriptor: | CHLORIDE ION, Isoaspartyl peptidase subunit alpha, Isoaspartyl peptidase subunit beta, ... | | Authors: | Sciuk, A, Jaskolski, M, Loch, J.I. | | Deposit date: | 2022-12-16 | | Release date: | 2023-05-03 | | Last modified: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The effects of nature-inspired amino acid substitutions on structural and biochemical properties of the E. coli L-asparaginase EcAIII.
Protein Sci., 32, 2023
|
|
8BP9
 
 | | Structure of E. coli Class 2 L-asparaginase EcAIII, mutant M200W (crystal M200W#2) | | Descriptor: | CHLORIDE ION, Isoaspartyl peptidase subunit alpha, Isoaspartyl peptidase subunit beta, ... | | Authors: | Sciuk, A, Jaskolski, M, Loch, J.I. | | Deposit date: | 2022-11-16 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The effects of nature-inspired amino acid substitutions on structural and biochemical properties of the E. coli L-asparaginase EcAIII.
Protein Sci., 32, 2023
|
|
3S85
 
 | | Discovery of New HIV Protease Inhibitors with Potential for Convenient Dosing and Reduced Side Effects: A-790742 and A-792611. | | Descriptor: | Protease/reverse transcriptase, methyl N-[(2S)-1-[[(2S,3S,5S)-5-[[(2S)-2-(methoxycarbonylamino)-3,3-dimethyl-butanoyl]amino]-3-oxidanyl-6-phenyl-1-(4-pyridin-3-ylphenyl)hexan-2-yl]amino]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | DeGoey, D.A, Flosi, W.J, Grampovnik, D.J, Flentge, C.A. | | Deposit date: | 2011-05-27 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2-Pyridyl P1'-substituted symmetry-based human immunodeficiency virus protease inhibitors (A-792611 and A-790742) with potential for convenient dosing and reduced side effects.
J.Med.Chem., 52, 2009
|
|
8SSH
 
 | | MtrR from Neisseria gonorrhoeae bound to Ethinyl Estradiol | | Descriptor: | Ethinyl estradiol, HTH-type transcriptional regulator MtrR, PHOSPHATE ION | | Authors: | Hooks, G.M, Brennan, R.G. | | Deposit date: | 2023-05-08 | | Release date: | 2024-02-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Hormonal steroids induce multidrug resistance and stress response genes in Neisseria gonorrhoeae by binding to MtrR.
Nat Commun, 15, 2024
|
|
3S41
 
 | | Glucokinase in complex with activator and glucose | | Descriptor: | Glucokinase, N,N-dimethyl-5-({2-methyl-6-[(5-methylpyrazin-2-yl)carbamoyl]-1-benzofuran-4-yl}oxy)pyrimidine-2-carboxamide, SODIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2011-05-18 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Designing glucokinase activators with reduced hypoglycemia risk: discovery of N,N-dimethyl-5-(2-methyl-6-((5-methylpyrazin-2-yl)-carbamoyl)benzofuran-4-yloxy)pyrimidine-2-carboxamide as a clinical candidate for the treatment of type 2 diabetes mellitus
MEDCHEMCOMM, 2, 2011
|
|
8CX4
 
 | | TCR-antigen complex AS8.4-YEIH-HLA*B27 | | Descriptor: | AS8.4a, AS8.4b, Beta-2-microglobulin, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
8DEW
 
 | |
8DEV
 
 | |
8DEU
 
 | |
3URF
 
 | | Human RANKL/OPG complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 11, soluble form, ... | | Authors: | Wang, X.Q, Luan, X.D, Lu, Q.Y. | | Deposit date: | 2011-11-22 | | Release date: | 2012-07-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystal Structure of Human RANKL Complexed with Its Decoy Receptor Osteoprotegerin
J.Immunol., 189, 2012
|
|
3V48
 
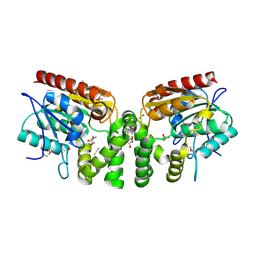 | | Crystal Structure of the putative alpha/beta hydrolase RutD from E.coli | | Descriptor: | GLYCEROL, Putative aminoacrylate hydrolase RutD, THIOCYANATE ION | | Authors: | Knapik, A.A, Petkowski, J.J, Otwinowski, Z, Cymborowski, M.T, Cooper, D.R, Chruszcz, M, Porebski, P.J, Niedzialkowska, E, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-04 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A multi-faceted analysis of RutD reveals a novel family of alpha / beta hydrolases.
Proteins, 80, 2012
|
|
3V4D
 
 | | Crystal structure of RutC protein a member of the YjgF family from E.coli | | Descriptor: | Aminoacrylate peracid reductase RutC | | Authors: | Knapik, A.A, Petkowski, J.J, Otwinowski, Z, Cymborowski, M.T, Cooper, D.R, Chruszcz, M, Porebski, P.J, Niedzialkowska, E, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-04 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Escherichia coli RutC, a member of the YjgF family and putative aminoacrylate peracid reductase of the rut operon.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3W9C
 
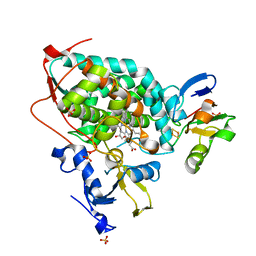 | | Crystal structure of the electron transfer complex of cytochrome p450cam with putidaredoxin | | Descriptor: | Camphor 5-monooxygenase, FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, ... | | Authors: | Kikui, Y, Hiruma, Y, Hass, M.A, Koteishi, H, Ubbink, M, Nojiri, M. | | Deposit date: | 2013-04-03 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of the cytochrome p450cam-putidaredoxin complex determined by paramagnetic NMR spectroscopy and crystallography.
J.Mol.Biol., 425, 2013
|
|
5G1Y
 
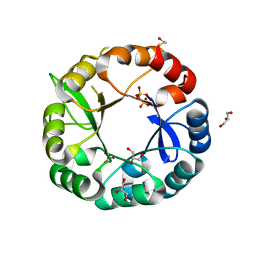 | | S. enterica HisA mutant D10G, dup13-15,V14:2M, Q24L, G102 | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, GLYCEROL, SULFATE ION | | Authors: | Guo, X, Soderholm, A, Newton, M, Nasvall, J, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2016-04-01 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4KR7
 
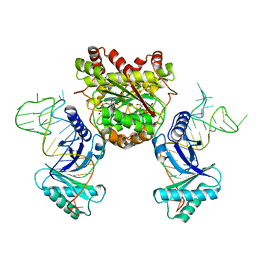 | | Crystal structure of a 4-thiouridine synthetase - RNA complex with bound ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Probable tRNA sulfurtransferase, ... | | Authors: | Neumann, P, Ficner, R, Lakomek, K. | | Deposit date: | 2013-05-16 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.421 Å) | | Cite: | Crystal structure of a 4-thiouridine synthetase-RNA complex reveals specificity of tRNA U8 modification.
Nucleic Acids Res., 42, 2014
|
|
4KR9
 
 | |
5G5J
 
 | | Crystal structure of human CYP3A4 bound to metformin | | Descriptor: | CYTOCHROME P450 3A4, Metformin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I. | | Deposit date: | 2016-05-25 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Heme Binding Biguanides Target Cytochrome P450-Dependent Cancer Cell Mitochondria.
Cell Chem Biol, 24, 2017
|
|
5GXG
 
 | | High-resolution crystal structure of the electron transfer complex of cytochrome p450cam with putidaredoxin | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Camphor 5-monooxygenase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Kikui, Y, Hiruma, Y, Ubbink, M, Nojiri, M. | | Deposit date: | 2016-09-17 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of productive and futile encounters in an electron transfer protein complex
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5G5I
 
 | | S. enterica HisA mutant D10G | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, PHOSPHATE ION | | Authors: | Guo, X, Soderholm, A, Newton, M, Nasvall, J, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2016-05-25 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5G2W
 
 | | S. enterica HisA mutant D10G, G102A | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, SULFATE ION | | Authors: | Guo, X, Soderholm, A, Newton, M, Nasvall, J, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2016-04-14 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5G4E
 
 | | S. enterica HisA mutant D10G, Dup13-15, Q24L, G102A, V106L | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, ACETIC ACID, GLYCEROL, ... | | Authors: | Soderholm, A, Guo, X, Newton, M, Nasvall, J, Duarte, F, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2016-05-12 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5G2I
 
 | | S. enterica HisA mutant Dup13-15(VVR) | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, PHOSPHATE ION, SODIUM ION | | Authors: | Guo, X, Soderholm, A, Newton, M, Nasvall, J, Duarte, F, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2016-04-08 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5G4W
 
 | | S. enterica HisA mutant D7N, D10G, Dup13-15 (VVR) with substrate ProFAR | | Descriptor: | GLYCEROL, HISA, [(2R,3S,4R,5R)-5-[4-aminocarbonyl-5-[(E)-[[(2R,3R,4S,5R)-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]amino]methylideneamino]imidazol-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Guo, X, Soderholm, A, Newton, M, Nasvall, J, Duarte, F, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2016-05-17 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1D70
 
 | |
5G1T
 
 | | S. enterica HisA mutant dup13-15, D10G | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, PHOSPHATE ION | | Authors: | Guo, X, Soderholm, A, Newton, M, Nasvall, J, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2016-03-30 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
