3VVT
 
 | | Crystal structure of reconstructed archaeal ancestral NDK, Arc1 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Nemoto, N, Miyazono, K, Kimura, M, Yokobori, S, Akanuma, S, Tanokura, M, Yamagishi, A. | | Deposit date: | 2012-07-27 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Experimental evidence for the thermophilicity of ancestral life
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3W21
 
 | | Crystal Structure of a Novel N-Substituted L-Amino Acid Dioxygenase in complex with alpha-KG from Burkholderia ambifaria AMMD | | Descriptor: | 2-OXOGLUTARIC ACID, Putative uncharacterized protein, ZINC ION | | Authors: | Qin, H.M, Miyakawa, T, Jia, M.Z, Nakamura, A, Ohtsuka, J, Xue, Y.L, Kawashima, T, Kasahara, T, Hibi, M, Ogawa, J, Tanokura, M. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of a Novel N-Substituted L-Amino Acid Dioxygenase from Burkholderia ambifaria AMMD
Plos One, 8, 2013
|
|
3WI7
 
 | | Crystal Structure of the Novel Haloalkane Dehalogenase DatA from Agrobacterium tumefaciens C58 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Haloalkane dehalogenase | | Authors: | Guan, L.J, Yabuki, H, Okai, M, Ohtsuka, J, Tanokura, M. | | Deposit date: | 2013-09-06 | | Release date: | 2014-07-23 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the novel haloalkane dehalogenase DatA from Agrobacterium tumefaciens C58 reveals a special halide-stabilizing pair and enantioselectivity mechanism.
Appl.Microbiol.Biotechnol., 98, 2014
|
|
3WG6
 
 | | Crystal structure of conjugated polyketone reductase C1 from Candida parapsilosis complexed with NADPH | | Descriptor: | Conjugated polyketone reductase C1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Qin, H.-M, Yamamura, A, Miyakawa, T, Maruoka, S, Ohtsuka, J, Nagata, K, Kataoka, M, Shimizu, S, Tanokura, M. | | Deposit date: | 2013-07-28 | | Release date: | 2013-08-21 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of conjugated polyketone reductase (CPR-C1) from Candida parapsilosis IFO 0708 complexed with NADPH.
Proteins, 81, 2013
|
|
3WIB
 
 | | Crystal structure of Y109W Mutant Haloalkane Dehalogenase DatA from Agrobacterium tumefaciens C58 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Haloalkane dehalogenase | | Authors: | Guan, L.J, Yabuki, H, Okai, M, Ohtsuka, J, Tanokura, M. | | Deposit date: | 2013-09-09 | | Release date: | 2014-07-23 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the novel haloalkane dehalogenase DatA from Agrobacterium tumefaciens C58 reveals a special halide-stabilizing pair and enantioselectivity mechanism.
Appl.Microbiol.Biotechnol., 98, 2014
|
|
3WAF
 
 | | X-ray structure of apo-TtFbpA, a ferric ion-binding protein from thermus thermophilus HB8 | | Descriptor: | ISOPROPYL ALCOHOL, Iron ABC transporter, periplasmic iron-binding protein | | Authors: | Wang, S, Ogata, M, Horita, S, Ohtsuka, J, Nagata, K, Tanokura, M. | | Deposit date: | 2013-05-02 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel mode of ferric ion coordination by the periplasmic ferric ion-binding subunit FbpA of an ABC-type iron transporter from Thermus thermophilus HB8.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WAE
 
 | | X-ray structure of Fe(III)-bicarbonates-ttfbpa, a ferric ion-binding protein from thermus thermophilus HB8 | | Descriptor: | BICARBONATE ION, FE (III) ION, IRON ABC TRANSPORTER, ... | | Authors: | Wang, S, Ogata, M, Horita, S, Ohtsuka, J, Nagata, K, Tanokura, M. | | Deposit date: | 2013-05-02 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A novel mode of ferric ion coordination by the periplasmic ferric ion-binding subunit FbpA of an ABC-type iron transporter from Thermus thermophilus HB8.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WWH
 
 | | Crystal structure of the first R-stereoselective -transaminase identified from Arthrobacter sp. KNK168 (FERM-BP-5228) | | Descriptor: | (R)-amine transaminase, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Guan, L.J, Ohtsuka, J, Okai, M, Miyakawa, T, Mase, T, Zhi, Y, Ito, N, Yasohara, Y, Tanokura, M. | | Deposit date: | 2014-06-18 | | Release date: | 2015-08-12 | | Last modified: | 2018-11-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A new target region for changing the substrate specificity of amine transaminases.
Sci Rep, 5, 2015
|
|
3WQB
 
 | | Crystal structure of aeromonas sobria serine protease (ASP) and the chaperone (ORF2) complex | | Descriptor: | CALCIUM ION, Extracellular serine protease, Open reading frame 2 | | Authors: | Kobayashi, H, Yoshida, T, Miyakawa, T, Kato, R, Tashiro, M, Yamanaka, H, Tanokura, M, Tsuge, H. | | Deposit date: | 2014-01-24 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural Basis for Action of the External Chaperone for a Propeptide-deficient Serine Protease from Aeromonas sobria.
J.Biol.Chem., 290, 2015
|
|
3WWJ
 
 | | Crystal structure of an engineered sitagliptin-producing transaminase, ATA-117-Rd11 | | Descriptor: | (R)-amine transaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Guan, L.J, Ohtsuka, J, Okai, M, Miyakawa, T, Mase, T, Zhi, Y, Hou, F, Ito, N, Yasohara, Y, Tanokura, M. | | Deposit date: | 2014-06-18 | | Release date: | 2015-08-12 | | Last modified: | 2018-11-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A new target region for changing the substrate specificity of amine transaminases.
Sci Rep, 5, 2015
|
|
1ZR7
 
 | |
2DE4
 
 | | Crystal structure of DSZB C27S mutant in complex with biphenyl-2-sulfinic acid | | Descriptor: | 1,1'-BIPHENYL-2-SULFINIC ACID, ACETATE ION, DIBENZOTHIOPHENE DESULFURIZATION ENZYME B | | Authors: | Lee, W.C, Ohshiro, T, Matsubara, T, Izumi, Y, Tanokura, M. | | Deposit date: | 2006-02-08 | | Release date: | 2006-08-01 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and desulfurization mechanism of 2'-hydroxybiphenyl-2-sulfinic acid desulfinase.
J.Biol.Chem., 281, 2006
|
|
2DE2
 
 | | Crystal structure of desulfurization enzyme DSZB | | Descriptor: | ACETATE ION, DIBENZOTHIOPHENE DESULFURIZATION ENZYME B, GLYCEROL | | Authors: | Lee, W.C, Ohshiro, T, Matsubara, T, Izumi, Y, Tanokura, M. | | Deposit date: | 2006-02-08 | | Release date: | 2006-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and desulfurization mechanism of 2'-hydroxybiphenyl-2-sulfinic acid desulfinase.
J.Biol.Chem., 281, 2006
|
|
2DE3
 
 | | Crystal structure of DSZB C27S mutant in complex with 2'-hydroxybiphenyl-2-sulfinic acid | | Descriptor: | 2'-HYDROXY-1,1'-BIPHENYL-2-SULFINIC ACID, ACETATE ION, DIBENZOTHIOPHENE DESULFURIZATION ENZYME B, ... | | Authors: | Lee, W.C, Ohshiro, T, Matsubara, T, Izumi, Y, Tanokura, M. | | Deposit date: | 2006-02-08 | | Release date: | 2006-08-01 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure and desulfurization mechanism of 2'-hydroxybiphenyl-2-sulfinic acid desulfinase.
J.Biol.Chem., 281, 2006
|
|
2EJX
 
 | | Crystal structure of the hypothetical protein STK_08120 from Sulfolobus tokodaii | | Descriptor: | STK_08120 | | Authors: | Miyakawa, T, Miyazono, K, Sawano, Y, Hatano, K, Nagata, K, Tanokura, M. | | Deposit date: | 2007-03-21 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A thermoacidophile-specific protein family, DUF3211, functions as a fatty acid carrier with novel binding mode
J.Bacteriol., 195, 2013
|
|
3WAZ
 
 | | Crystal structure of a restriction enzyme PabI in complex with DNA | | Descriptor: | ADENINE, DNA (5'-D(*GP*CP*AP*TP*AP*GP*CP*TP*GP*TP*(ORP)P*CP*AP*GP*CP*TP*AP*TP*GP*C)-3'), Putative uncharacterized protein | | Authors: | Miyazono, K, Miyakawa, T, Ito, T, Tanokura, M. | | Deposit date: | 2013-05-10 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A sequence-specific DNA glycosylase mediates restriction-modification in Pyrococcus abyssi.
Nat Commun, 5, 2014
|
|
3WWI
 
 | | Crystal structure of the G136F mutant of the first R-stereoselective -transaminase identified from Arthrobacter sp. KNK168 (FERM-BP-5228) | | Descriptor: | (R)-amine transaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Guan, L.J, Ohtsuka, J, Miyakawa, T, Zhi, Y, Ito, N, Yasohara, Y, Tanokura, M. | | Deposit date: | 2014-06-18 | | Release date: | 2015-08-19 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | A new target region for changing the substrate specificity of amine transaminases.
Sci Rep, 5, 2015
|
|
3WDS
 
 | |
2EB1
 
 | | Crystal Structure of the C-Terminal RNase III Domain of Human Dicer | | Descriptor: | Endoribonuclease Dicer, MAGNESIUM ION | | Authors: | Takeshita, D, Zenno, S, Lee, W.C, Nagata, K, Saigo, K, Tanokura, M. | | Deposit date: | 2007-02-05 | | Release date: | 2007-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homodimeric Structure and Double-stranded RNA Cleavage Activity of the C-terminal RNase III Domain of Human Dicer
J.Mol.Biol., 374, 2007
|
|
2DYF
 
 | |
3TZ1
 
 | | Crystal structure of the Ca2+-saturated C-terminal domain of Akazara scallop troponin C in complex with a troponin I fragment | | Descriptor: | CALCIUM ION, Troponin C, Troponin I | | Authors: | Yumoto, F, Kato, Y.S, Ohtsuki, I, Tanokura, M. | | Deposit date: | 2011-09-26 | | Release date: | 2013-01-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Ca2+-saturated C-terminal domain of scallop troponin C in complex with a troponin I fragment
Biol.Chem., 394, 2012
|
|
3VS9
 
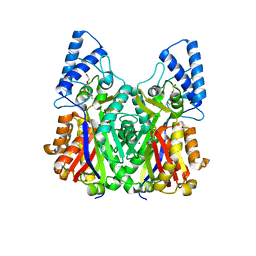 | | Crystal structure of type III PKS ArsC mutant | | Descriptor: | SODIUM ION, TETRAETHYLENE GLYCOL, Type III polyketide synthase | | Authors: | Satou, R, Miyanaga, A, Ozawa, H, Funa, N, Miyazono, K, Tanokura, M, Ohnishi, Y, Horinouchi, S. | | Deposit date: | 2012-04-23 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis for cyclization specificity of two Azotobacter type III polyketide synthases: a single amino acid substitution reverses their cyclization specificity
J.Biol.Chem., 288, 2013
|
|
3VGK
 
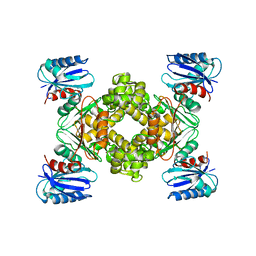 | | Crystal structure of a ROK family glucokinase from Streptomyces griseus | | Descriptor: | Glucokinase, SULFATE ION, ZINC ION | | Authors: | Miyazono, K, Tabei, N, Morita, S, Ohnishi, Y, Horinouchi, S, Tanokura, M. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Substrate recognition mechanism and substrate-dependent conformational changes of an ROK family glucokinase from Streptomyces griseus
J.Bacteriol., 194, 2012
|
|
3VS8
 
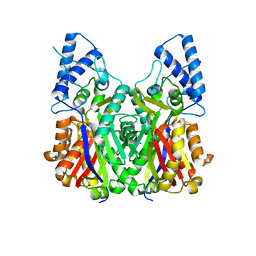 | | Crystal structure of type III PKS ArsC | | Descriptor: | SODIUM ION, Type III polyketide synthase | | Authors: | Satou, R, Miyanaga, A, Ozawa, H, Funa, N, Miyazono, K, Tanokura, M, Ohnishi, Y, Horinouchi, S. | | Deposit date: | 2012-04-23 | | Release date: | 2013-04-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural basis for cyclization specificity of two Azotobacter type III polyketide synthases: a single amino acid substitution reverses their cyclization specificity
J.Biol.Chem., 288, 2013
|
|
3VTG
 
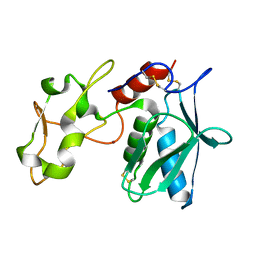 | | High choriolytic enzyme 1 (HCE-1), a hatching enzyme zinc-protease from Oryzias latipes (Medaka fish) | | Descriptor: | High choriolytic enzyme 1, ZINC ION | | Authors: | Kudo, N, Yasumasu, S, Iuchi, I, Tanokura, M. | | Deposit date: | 2012-05-30 | | Release date: | 2013-06-05 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal Structure of High choriolytic enzyme 1 (HCE-1), a hatching enzyme from Oryzias latipes (Medaka fish)
To be Published
|
|
