1OFF
 
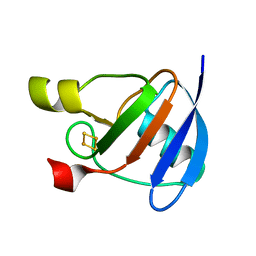 | |
1JQT
 
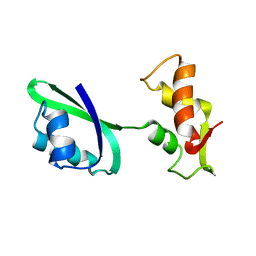 | | Fitting of L11 protein in the low resolution cryo-EM map of E.coli 70S ribosome | | Descriptor: | 50S Ribosomal protein L11 | | Authors: | Agrawal, R.K, Linde, J, Segupta, J, Nierhaus, K.H, Frank, J. | | Deposit date: | 2001-08-07 | | Release date: | 2001-09-07 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation.
J.Mol.Biol., 311, 2001
|
|
2I5P
 
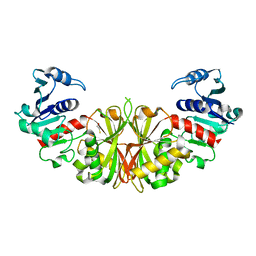 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase isoform 1 from K. marxianus | | Descriptor: | BETA-MERCAPTOETHANOL, Glyceraldehyde-3-phosphate dehydrogenase 1, alpha-D-glucopyranose | | Authors: | Ferreira-da-Silva, F, Pereira, P.J.B, Gales, L, Moradas-Ferreira, P, Damas, A.M. | | Deposit date: | 2006-08-25 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal and Solution Structures of Glyceraldehyde-3-phosphate Dehydrogenase Reveal Different Quaternary Structures.
J.Biol.Chem., 281, 2006
|
|
4V48
 
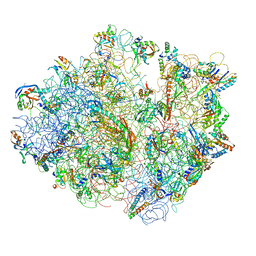 | | Real space refined coordinates of the 30S and 50S subunits fitted into the low resolution cryo-EM map of the initiation-like state of E. coli 70S ribosome | | Descriptor: | 16S RIBOSOMAL RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, H, Sengupta, J, Valle, M, Korostelev, A, Eswar, N, Stagg, S.M, Van Roey, P, Agrawal, R.K, Harvey, S.T, Sali, A, Chapman, M.S, Frank, J. | | Deposit date: | 2003-05-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Study of the structural dynamics of the E. coli 70S ribosome using real space refinement
Cell(Cambridge,Mass.), 113, 2003
|
|
2IHR
 
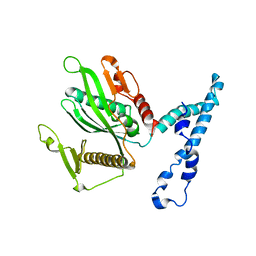 | | RF2 of Thermus thermophilus | | Descriptor: | Peptide chain release factor 2 | | Authors: | Dobbek, H, Voertler, C.S, Sprinzl, M. | | Deposit date: | 2006-09-27 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Release factors 2 from Escherichia coli and Thermus thermophilus: structural, spectroscopic and microcalorimetric studies.
Nucleic Acids Res., 35, 2007
|
|
4V47
 
 | | Real space refined coordinates of the 30S and 50S subunits fitted into the low resolution cryo-EM map of the EF-G.GTP state of E. coli 70S ribosome | | Descriptor: | 16S RIBOSOMAL RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, H, Sengupta, J, Valle, M, Korostelev, A, Eswar, N, Stagg, S.M, Van Roey, P, Agrawal, R.K, Harvey, S.T, Sali, A, Chapman, M.S, Frank, J. | | Deposit date: | 2003-05-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (12.3 Å) | | Cite: | Study of the structural dynamics of the E. coli 70S ribosome using real space refinement
Cell(Cambridge,Mass.), 113, 2003
|
|
2JXY
 
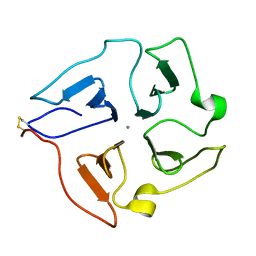 | | Solution structure of the hemopexin-like domain of MMP12 | | Descriptor: | CALCIUM ION, Macrophage metalloelastase | | Authors: | Bertini, I, Calderone, V, Fragai, M, Jaiswal, R, Luchinat, C, Melikian, M. | | Deposit date: | 2007-12-01 | | Release date: | 2008-05-27 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Evidence of reciprocal reorientation of the catalytic and hemopexin-like domains of full-length MMP-12
J.Am.Chem.Soc., 130, 2008
|
|
2JB6
 
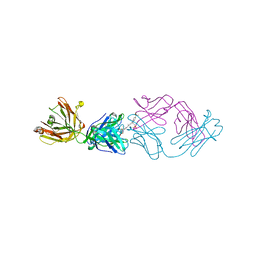 | | Fab fragment in complex with small molecule hapten, crystal form-2 | | Descriptor: | 2-{(1E,3Z,5E,7E)-7-[3,3-DIMETHYL-5-SULFO-1-(2-SULFOETHYL)-1,3-DIHYDRO-2H-INDOL-2-YLIDENE]-4-METHYLHEPTA-1,3,5-TRIEN-1-YL}-3,3-DIMETHYL-5-SULFO-1-(2-SULFOETHYL)-3H-INDOLIUM, FAB FRAGMENT MOR03268 HEAVY CHAIN, FAB FRAGMENT MOR03268 LIGHT CHAIN | | Authors: | Hillig, R.C, Baesler, S, Malawski, G, Badock, V, Bahr, I, Schirner, M, Licha, K. | | Deposit date: | 2006-12-03 | | Release date: | 2008-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Fab Mor03268 Triggers Absorption Shift of a Diagnostic Dye Via Packaging in a Solvent-Shielded Fab Dimer Interface
J.Mol.Biol., 377, 2008
|
|
2JB5
 
 | | Fab fragment in complex with small molecule hapten, crystal form-1 | | Descriptor: | 2-{(1E,3Z,5E,7E)-7-[3,3-DIMETHYL-5-SULFO-1-(2-SULFOETHYL)-1,3-DIHYDRO-2H-INDOL-2-YLIDENE]-4-METHYLHEPTA-1,3,5-TRIEN-1-YL}-3,3-DIMETHYL-5-SULFO-1-(2-SULFOETHYL)-3H-INDOLIUM, FAB FRAGMENT MOR03268 HEAVY CHAIN, FAB FRAGMENT MOR03268 LIGHT CHAIN | | Authors: | Hillig, R.C, Baesler, S, Malawski, G, Badock, V, Bahr, I, Schirner, M, Licha, K. | | Deposit date: | 2006-12-03 | | Release date: | 2008-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fab Mor03268 Triggers Absorption Shift of a Diagnostic Dye Via Packaging in a Solvent-Shielded Fab Dimer Interface
J.Mol.Biol., 377, 2008
|
|
2K7Q
 
 | | Filamin A Ig-like domains 18-19 | | Descriptor: | Filamin-A | | Authors: | Heikkinen, O.K, Kilpelainen, I, Koskela, H, Permi, P, Heikkinen, S, Ylanne, J. | | Deposit date: | 2008-08-19 | | Release date: | 2009-07-07 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Atomic structures of two novel immunoglobulin-like domain pairs in the actin cross-linking protein filamin
J.Biol.Chem., 284, 2009
|
|
2K7P
 
 | | Filamin A Ig-like domains 16-17 | | Descriptor: | Filamin-A | | Authors: | Heikkinen, O.K, Kilpelainen, I, Koskela, H, Permi, P, Heikkinen, S, Ylanne, J. | | Deposit date: | 2008-08-19 | | Release date: | 2009-07-07 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Atomic structures of two novel immunoglobulin-like domain pairs in the actin cross-linking protein filamin
J.Biol.Chem., 284, 2009
|
|
2O0T
 
 | |
3O08
 
 | | Crystal structure of dimeric KlHxk1 in crystal form I | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hexokinase, SULFATE ION | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-07-19 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O4W
 
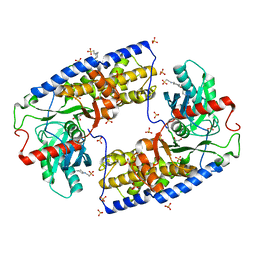 | | Crystal structure of dimeric KlHxk1 in crystal form IV | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Hexokinase, ... | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-07-27 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O8M
 
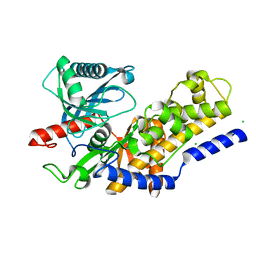 | | Crystal structure of monomeric KlHxk1 in crystal form XI with glucose bound (closed state) | | Descriptor: | CHLORIDE ION, Hexokinase, alpha-D-glucopyranose, ... | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-08-03 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O80
 
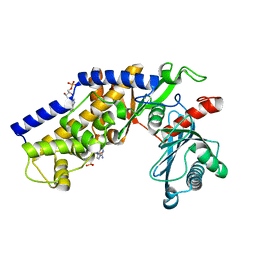 | | Crystal structure of monomeric KlHxk1 in crystal form IX (open state) | | Descriptor: | Hexokinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-08-02 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3OEQ
 
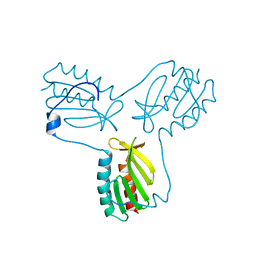 | | Crystal structure of trimeric frataxin from the yeast Saccharomyces cerevisiae, with full length n-terminus | | Descriptor: | Frataxin homolog, mitochondrial | | Authors: | Soderberg, C.A.G, Rajan, S, Gakh, O, Ta, C, Isaya, G, Al-Karadaghi, S. | | Deposit date: | 2010-08-13 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Oligomerization Propensity and Flexibility of Yeast Frataxin Studied by X-ray Crystallography and Small-Angle X-ray Scattering.
J.Mol.Biol., 414, 2011
|
|
3OER
 
 | | Crystal structure of trimeric frataxin from the yeast saccharomyces cerevisiae, complexed with cobalt | | Descriptor: | COBALT (II) ION, Frataxin homolog, mitochondrial | | Authors: | Soderberg, C.A.G, Rajan, S, Gakh, O, Ta, C, Isaya, G, Al-Karadaghi, S. | | Deposit date: | 2010-08-13 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Oligomerization Propensity and Flexibility of Yeast Frataxin Studied by X-ray Crystallography and Small-Angle X-ray Scattering.
J.Mol.Biol., 414, 2011
|
|
3R3L
 
 | | Structure of NP protein from Lassa AV strain | | Descriptor: | MANGANESE (II) ION, Nucleoprotein, ZINC ION | | Authors: | Perbandt, M, Brunotte, L, Gunther, S, Betzel, C. | | Deposit date: | 2011-03-16 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Structure of the Lassa virus nucleoprotein revealed by X-ray crystallography, small-angle X-ray scattering, and electron microscopy.
J.Biol.Chem., 286, 2011
|
|
3RBS
 
 | |
3RQC
 
 | | Crystal structure of the catalytic core of the 2-oxoacid dehydrogenase multienzyme complex from Thermoplasma acidophilum | | Descriptor: | Probable lipoamide acyltransferase | | Authors: | Marrott, N.L, Crennell, S.J, Hough, D.W, Danson, M.J, van den Elsen, J.M.H. | | Deposit date: | 2011-04-28 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.01 Å) | | Cite: | The catalytic core of an archaeal 2-oxoacid dehydrogenase multienzyme complex is a 42-mer protein assembly.
Febs J., 279, 2012
|
|
3STQ
 
 | |
3SW0
 
 | | Structure of the C-terminal region (modules 18-20) of complement regulator Factor H | | Descriptor: | Complement factor H, GLYCEROL, PHOSPHATE ION | | Authors: | Morgan, H.P, Guariento, M, Schmidt, C.Q, Barlow, P.N, Hannan, J.P. | | Deposit date: | 2011-07-13 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of the C-Terminal Region (Modules 18-20) of Complement Regulator Factor H (FH).
Plos One, 7, 2012
|
|
3T06
 
 | |
4C9T
 
 | | BACTERIAL CHALCONE ISOMERASE IN open CONFORMATION FROM EUBACTERIUM RAMULUS AT 2.0 A RESOLUTION, SelenoMet derivative | | Descriptor: | CHALCONE ISOMERASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thomsen, M, Palm, G.J, Hinrichs, W. | | Deposit date: | 2013-10-03 | | Release date: | 2014-10-22 | | Last modified: | 2015-04-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and Catalytic Mechanism of the Evolutionarily Unique Bacterial Chalcone Isomerase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
