8HWK
 
 | | Limosilactobacillus reuteri N1 GtfB-maltohexaose | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Dong, J.J, Bai, Y.X. | | Deposit date: | 2022-12-30 | | Release date: | 2024-01-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights into the Structure-Function Relationship of GH70 GtfB alpha-Glucanotransferases from the Crystal Structure and Molecular Dynamic Simulation of a Newly Characterized Limosilactobacillus reuteri N1 GtfB Enzyme.
J.Agric.Food Chem., 72, 2024
|
|
8HU4
 
 | | Limosilactobacillus reuteri N1 GtfB | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Dong, J.J, Bai, Y.X. | | Deposit date: | 2022-12-22 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Insights into the Structure-Function Relationship of GH70 GtfB alpha-Glucanotransferases from the Crystal Structure and Molecular Dynamic Simulation of a Newly Characterized Limosilactobacillus reuteri N1 GtfB Enzyme.
J.Agric.Food Chem., 72, 2024
|
|
1HT6
 
 | | CRYSTAL STRUCTURE AT 1.5A RESOLUTION OF THE BARLEY ALPHA-AMYLASE ISOZYME 1 | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-AMYLASE ISOZYME 1, CALCIUM ION | | Authors: | Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2000-12-29 | | Release date: | 2003-07-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of barley alpha-amylase isozyme 1 reveals a novel role of domain C in substrate recognition and binding: a pair of sugar tongs
Structure, 11, 2003
|
|
7E33
 
 | | Serotonin 1E (5-HT1E) receptor-Gi protein complex | | Descriptor: | 3-(1-methylpiperidin-4-yl)-1H-indol-5-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-14 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
7E2X
 
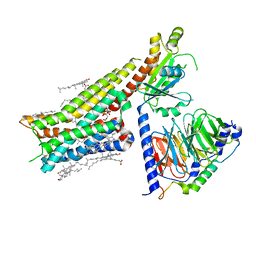 | | Apo serotonin 1A (5-HT1A) receptor-Gi protein complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-14 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
7E2Y
 
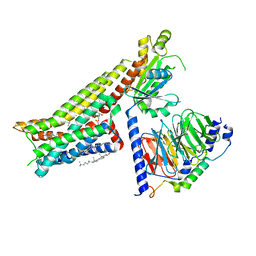 | | Serotonin-bound Serotonin 1A (5-HT1A) receptor-Gi protein complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-14 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
7E2Z
 
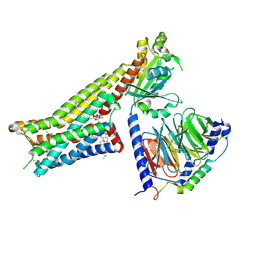 | | Aripiprazole-bound serotonin 1A (5-HT1A) receptor-Gi protein complex | | Descriptor: | 7-[4-[4-[2,3-bis(chloranyl)phenyl]piperazin-1-yl]butoxy]-3,4-dihydro-1H-quinolin-2-one, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-14 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
7E32
 
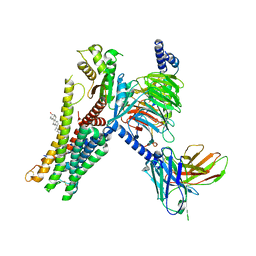 | | Serotonin 1D (5-HT1D) receptor-Gi protein complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-21 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
2PRM
 
 | | The structures of apo- and inhibitor bound human dihydroorotate dehydrogenase reveal conformational flexibility within the inhibitor binding site | | Descriptor: | Dihydroorotate dehydrogenase, mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Walse, B, Dufe, V.T, Al-Karadaghi, S. | | Deposit date: | 2007-05-04 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structures of human dihydroorotate dehydrogenase with and without inhibitor reveal conformational flexibility in the inhibitor and substrate binding sites
Biochemistry, 47, 2008
|
|
4ZS9
 
 | | Raffinose and panose binding protein from Bifidobacterium animalis subsp. lactis Bl-04, bound with raffinose | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Sugar binding protein of ABC transporter system, ... | | Authors: | Fredslund, F, Ejby, M, Andersen, J.M, Slotboom, D.J, Abou Hachem, M. | | Deposit date: | 2015-05-13 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | An ATP Binding Cassette Transporter Mediates the Uptake of alpha-(1,6)-Linked Dietary Oligosaccharides in Bifidobacterium and Correlates with Competitive Growth on These Substrates.
J. Biol. Chem., 291, 2016
|
|
2PRL
 
 | | The structures of apo- and inhibitor bound human dihydroorotate dehydrogenase reveal conformational flexibility within the inhibitor binding site | | Descriptor: | 5-METHOXY-2-[(4-PHENOXYPHENYL)AMINO]BENZOIC ACID, ACETATE ION, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, ... | | Authors: | Walse, B, Dufe, V.T, Al-Karadaghi, S. | | Deposit date: | 2007-05-04 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structures of human dihydroorotate dehydrogenase with and without inhibitor reveal conformational flexibility in the inhibitor and substrate binding sites
Biochemistry, 47, 2008
|
|
4ZZE
 
 | | Raffinose and panose binding protein from Bifidobacterium animalis subsp. lactis Bl-04, bound with panose | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Sugar binding protein of ABC transporter system, ... | | Authors: | Fredslund, F, Ejby, M, Andersen, J.M, Slotboom, D.J, Hachem, M.A. | | Deposit date: | 2015-05-22 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An ATP Binding Cassette Transporter Mediates the Uptake of alpha-(1,6)-Linked Dietary Oligosaccharides in Bifidobacterium and Correlates with Competitive Growth on These Substrates.
J. Biol. Chem., 291, 2016
|
|
4ZZA
 
 | | Raffinose and panose binding protein from Bifidobacterium animalis subsp. lactis Bl-04, bound with raffinose, selenomethionine derivative | | Descriptor: | Sugar binding protein of ABC transporter system, alpha-D-galactopyranose-(1-6)-alpha-D-glucopyranose-(1-2)-beta-D-fructofuranose | | Authors: | Fredslund, F, Ejby, M, Andersen, J.M, Slotboom, D.J, Abou Hachem, M. | | Deposit date: | 2015-05-22 | | Release date: | 2016-06-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | An ATP Binding Cassette Transporter Mediates the Uptake of alpha-(1,6)-Linked Dietary Oligosaccharides in Bifidobacterium and Correlates with Competitive Growth on These Substrates.
J. Biol. Chem., 291, 2016
|
|
2PRH
 
 | | The structures of apo- and inhibitor bound human dihydroorotate dehydrogenase reveal conformational flexibility within the inhibitor binding site | | Descriptor: | 6-CHLORO-2-(2'-FLUOROBIPHENYL-4-YL)-3-METHYLQUINOLINE-4-CARBOXYLIC ACID, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, Dihydroorotate dehydrogenase, ... | | Authors: | Walse, B, Dufe, V.T, Al-Karadaghi, S. | | Deposit date: | 2007-05-04 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structures of human dihydroorotate dehydrogenase with and without inhibitor reveal conformational flexibility in the inhibitor and substrate binding sites
Biochemistry, 47, 2008
|
|
8BDD
 
 | |
6Y9T
 
 | | Family GH13_31 enzyme | | Descriptor: | Alpha-glucosidase, CALCIUM ION | | Authors: | Andersen, S, Poulsen, J.C.N, Moeller, M.S, Abou Hachem, M, Lo Leggio, L. | | Deposit date: | 2020-03-10 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | An 1,4-alpha-Glucosyltransferase Defines a New Maltodextrin Catabolism Scheme in Lactobacillus acidophilus.
Appl.Environ.Microbiol., 86, 2020
|
|
8BDQ
 
 | |
6RZN
 
 | |
6RZO
 
 | |
2QPS
 
 | | "Sugar tongs" mutant Y380A in complex with acarbose | | Descriptor: | Alpha-amylase type A isozyme, CALCIUM ION | | Authors: | Aghajari, N, Jensen, M.H, Tranier, S, Haser, R. | | Deposit date: | 2007-07-25 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 'pair of sugar tongs' site on the non-catalytic domain C of barley alpha-amylase participates in substrate binding and activity
Febs J., 274, 2007
|
|
2QPU
 
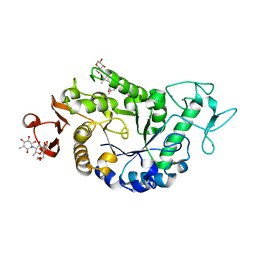 | | Sugar tongs mutant S378P in complex with acarbose | | Descriptor: | 1,2-ETHANEDIOL, 1,5-anhydro-4-O-(4,6-dideoxy-4-{[(1S,2S,3S,4R,5S,6R)-2,3,4,6-tetrahydroxy-5-methylcyclohexyl]amino}-alpha-D-glucopyranosyl)-D-glucitol, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Aghajari, N, Jensen, M.H, Tranier, S, Haser, R. | | Deposit date: | 2007-07-25 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 'pair of sugar tongs' site on the non-catalytic domain C of barley alpha-amylase participates in substrate binding and activity
Febs J., 274, 2007
|
|
5LRA
 
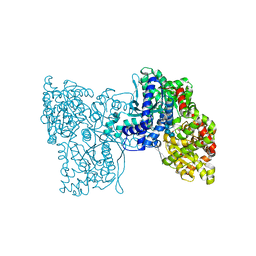 | | Plastidial phosphorylase PhoI from barley in complex with maltotetraose | | Descriptor: | Alpha-1,4 glucan phosphorylase, PYRIDOXAL-5'-PHOSPHATE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Cuesta-Seijo, J.A, Ruzanski, C, Krucewicz, K, Palcic, M.M. | | Deposit date: | 2016-08-18 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Functional and structural characterization of plastidic starch phosphorylase during barley endosperm development.
PLoS ONE, 12, 2017
|
|
5LRB
 
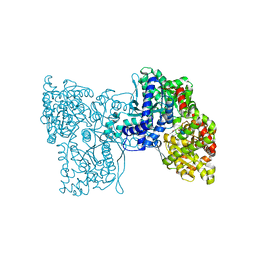 | | Plastidial phosphorylase from Barley in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-1,4 glucan phosphorylase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Cuesta-Seijo, J.A, Ruzanski, C, Kruzewicz, K, Palcic, M.M. | | Deposit date: | 2016-08-18 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Functional and structural characterization of plastidic starch phosphorylase during barley endosperm development.
PLoS ONE, 12, 2017
|
|
5LR8
 
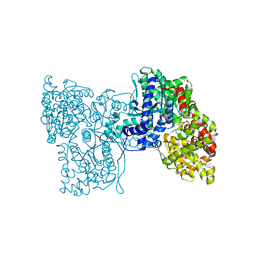 | | Structure of plastidial phosphorylase Pho1 from Barley | | Descriptor: | Alpha-1,4 glucan phosphorylase, CITRIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Cuesta-Seijo, J.A, Ruzanski, C, Kruzewicz, K, Palcic, M.M. | | Deposit date: | 2016-08-18 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Functional and structural characterization of plastidic starch phosphorylase during barley endosperm development.
PLoS ONE, 12, 2017
|
|
4AIE
 
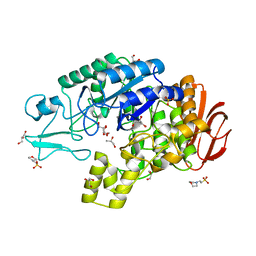 | | Structure of glucan-1,6-alpha-glucosidase from Lactobacillus acidophilus NCFM | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLUCAN 1,6-ALPHA-GLUCOSIDASE, ... | | Authors: | Fredslund, F, Navarro Poulsen, J.C, Lo Leggio, L. | | Deposit date: | 2012-02-09 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Enzymology and Structure of the Gh13_31 Glucan 1,6-Alpha-Glucosidase that Confers Isomaltooligosaccharide Utilization in the Probiotic Lactobacillus Acidophilus Ncfm.
J.Bacteriol., 194, 2012
|
|
