7K4X
 
 | | Crystal structure of Kemp Eliminase HG3.7 in complex with the transition state analog 6-nitrobenzotriazole | | 分子名称: | 6-NITROBENZOTRIAZOLE, ACETATE ION, Endo-1,4-beta-xylanase, ... | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4Y
 
 | | Crystal structure of Kemp Eliminase HG3.17 at 343 K | | 分子名称: | Endo-1,4-beta-xylanase | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4R
 
 | | Crystal structure of Kemp Eliminase HG3 K50Q | | 分子名称: | Endo-1,4-beta-xylanase | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4P
 
 | | Crystal structure of Kemp Eliminase HG3 | | 分子名称: | Endo-1,4-beta-xylanase | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4T
 
 | | Crystal structure of Kemp Eliminase HG3.17 | | 分子名称: | Endo-1,4-beta-xylanase | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (0.999 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4S
 
 | | Crystal structure of Kemp Eliminase HG3.7 | | 分子名称: | Endo-1,4-beta-xylanase, SULFATE ION | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4W
 
 | | Crystal structure of Kemp Eliminase HG3.17 in the inactive state | | 分子名称: | CALCIUM ION, Endo-1,4-beta-xylanase | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4Z
 
 | | Crystal structure of Kemp Eliminase HG3.17 in complex with the transition state analog 6-nitrobenzotriazole | | 分子名称: | 6-NITROBENZOTRIAZOLE, Endo-1,4-beta-xylanase, PENTAETHYLENE GLYCOL | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4Q
 
 | | Crystal structure of Kemp Eliminase HG3 in complex with the transition state analog 6-nitrobenzotriazole | | 分子名称: | 6-NITROBENZOTRIAZOLE, Endo-1,4-beta-xylanase, SULFATE ION | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4V
 
 | | Crystal structure of Kemp Eliminase HG3.17 | | 分子名称: | Endo-1,4-beta-xylanase, TETRAETHYLENE GLYCOL | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4U
 
 | | Crystal structure of Kemp Eliminase HG3 K50Q in complex with the transition state analog 6-nitrobenzotriazole | | 分子名称: | 6-NITROBENZOTRIAZOLE, Endo-1,4-beta-xylanase | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
6AID
 
 | | Structural insights into the unique polylactate degrading mechanism of Thermobifida alba cutinase | | 分子名称: | CALCIUM ION, Esterase, LACTIC ACID, ... | | 著者 | Kitadokoro, K, Kakara, M, Matsui, S, Osokoshi, R, Thumarat, U, Kawai, F, Kamitani, S. | | 登録日 | 2018-08-22 | | 公開日 | 2019-02-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structural insights into the unique polylactate-degrading mechanism of Thermobifida alba cutinase.
Febs J., 286, 2019
|
|
1HE8
 
 | | Ras G12V - PI 3-kinase gamma complex | | 分子名称: | MAGNESIUM ION, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT, GAMMA ISOFORM, ... | | 著者 | Pacold, M.E, Suire, S, Perisic, O, Lara-Gonzalez, S, Davis, C.T, Hawkins, P.T, Walker, E.H, Stephens, L, Eccleston, J.F, Williams, R.L. | | 登録日 | 2000-11-20 | | 公開日 | 2001-01-08 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal Structure and Functional Analysis of Ras Binding to its Effector Phosphoinositide 3-Kinase Gamma
Cell(Cambridge,Mass.), 103, 2000
|
|
6JLU
 
 | | Structure of PSII-FCP supercomplex from a centric diatom Chaetoceros gracilis at 3.02 angstrom resolution | | 分子名称: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | 著者 | Pi, X, Zhao, S, Wang, W, Kuang, T, Sui, S, Shen, J. | | 登録日 | 2019-03-06 | | 公開日 | 2019-07-31 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.02 Å) | | 主引用文献 | The pigment-protein network of a diatom photosystem II-light-harvesting antenna supercomplex.
Science, 365, 2019
|
|
5XMI
 
 | | Cryo-EM Structure of the ATP-bound VPS4 mutant-E233Q hexamer (masked) | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Vacuolar protein sorting-associated protein 4 | | 著者 | Sun, S, Li, L, Yang, F, Wang, X, Fan, F, Li, X, Wang, H, Sui, S. | | 登録日 | 2017-05-15 | | 公開日 | 2017-08-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM structures of the ATP-bound Vps4(E233Q) hexamer and its complex with Vta1 at near-atomic resolution
Nat Commun, 8, 2017
|
|
5XMK
 
 | | Cryo-EM structure of the ATP-bound Vps4 mutant-E233Q complex with Vta1 (masked) | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Vacuolar protein sorting-associated protein 4, Vacuolar protein sorting-associated protein VTA1 | | 著者 | Sun, S, Li, L, Yang, F, Wang, X, Fan, F, Li, X, Wang, H, Sui, S. | | 登録日 | 2017-05-15 | | 公開日 | 2017-08-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.18 Å) | | 主引用文献 | Cryo-EM structures of the ATP-bound Vps4(E233Q) hexamer and its complex with Vta1 at near-atomic resolution
Nat Commun, 8, 2017
|
|
7DF8
 
 | | full length hNPC1L1-Apo | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Hu, M, Sun, S, Sui, S. | | 登録日 | 2020-11-06 | | 公開日 | 2021-08-04 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | Structural insights into the mechanism of human NPC1L1-mediated cholesterol uptake.
Sci Adv, 7, 2021
|
|
6JIB
 
 | | Human MTHFD2 in complex with DS44960156 | | 分子名称: | 4-(5-oxo-1,5-dihydro-2H-[1]benzopyrano[3,4-c]pyridine-3(4H)-carbonyl)benzoic acid, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial, ... | | 著者 | Suzuki, M, Matsui, Y, Kawai, J. | | 登録日 | 2019-02-20 | | 公開日 | 2019-06-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structure-Based Design and Synthesis of an Isozyme-Selective MTHFD2 Inhibitor with a Tricyclic Coumarin Scaffold.
Acs Med.Chem.Lett., 10, 2019
|
|
6JID
 
 | | Human MTHFD2 in complex with Compound 1 | | 分子名称: | 5-(4-oxo-2-phenyl-1,5,7,8-tetrahydropyrido[4,3-d]pyrimidine-6(4H)-carbonyl)-1,3-dihydro-2H-2lambda~6~,1-benzothiazole-2,2-dione, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial, ... | | 著者 | Suzuki, M, Matsui, Y, Matsuhashi, N, Kawai, J. | | 登録日 | 2019-02-20 | | 公開日 | 2019-06-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure-Based Design and Synthesis of an Isozyme-Selective MTHFD2 Inhibitor with a Tricyclic Coumarin Scaffold.
Acs Med.Chem.Lett., 10, 2019
|
|
6KG2
 
 | | Human MTHFD2 in complex with Compound 18 | | 分子名称: | Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial, N-[2-chloranyl-4-[[7-methyl-8-(4-methylpiperazin-1-yl)-5-oxidanylidene-2,4-dihydro-1H-chromeno[3,4-c]pyridin-3-yl]carbonyl]phenyl]methanesulfonamide, ... | | 著者 | Suzuki, M, Matsui, Y, Ota, M, Kawai, J. | | 登録日 | 2019-07-10 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Discovery of a Potent, Selective, and Orally Available MTHFD2 Inhibitor (DS18561882) with in Vivo Antitumor Activity.
J.Med.Chem., 62, 2019
|
|
6XF4
 
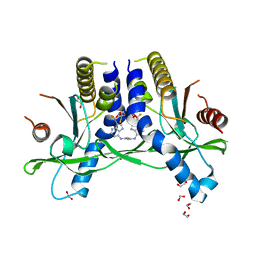 | | Crystal structure of STING REF variant in complex with E7766 | | 分子名称: | (1R,3R,15E,28R,29R,30R,31R,34R,36R,39S,41R)-29,41-difluoro-34,39-disulfanyl-2,33,35,38,40,42-hexaoxa-4,6,9,11,13,18,20,22,25,27-decaaza-34,39-diphosphaoctacyclo[28.6.4.1~3,36~.1~28,31~.0~4,8~.0~7,12~.0~19,24~.0~23,27~]dotetraconta-5,7,9,11,15,19,21,23,25-nonaene 34,39-dioxide (non-preferred name), 1,2-ETHANEDIOL, Stimulator of interferon genes protein | | 著者 | Chen, Y, Wang, J.Y, Kim, D.-S. | | 登録日 | 2020-06-15 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.77 Å) | | 主引用文献 | E7766, a Macrocycle-Bridged Stimulator of Interferon Genes (STING) Agonist with Potent Pan-Genotypic Activity.
Chemmedchem, 16, 2021
|
|
6XF3
 
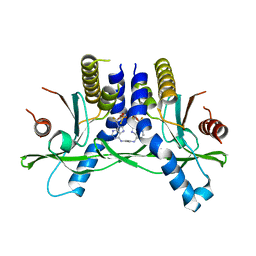 | | Crystal structure of STING in complex with E7766 | | 分子名称: | (1R,3R,15E,28R,29R,30R,31R,34R,36R,39S,41R)-29,41-difluoro-34,39-disulfanyl-2,33,35,38,40,42-hexaoxa-4,6,9,11,13,18,20,22,25,27-decaaza-34,39-diphosphaoctacyclo[28.6.4.1~3,36~.1~28,31~.0~4,8~.0~7,12~.0~19,24~.0~23,27~]dotetraconta-5,7,9,11,15,19,21,23,25-nonaene 34,39-dioxide (non-preferred name), Stimulator of interferon genes protein | | 著者 | Chen, Y, Wang, J.Y, Kim, D.-S. | | 登録日 | 2020-06-15 | | 公開日 | 2021-02-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | E7766, a Macrocycle-Bridged Stimulator of Interferon Genes (STING) Agonist with Potent Pan-Genotypic Activity.
Chemmedchem, 16, 2021
|
|
5XWM
 
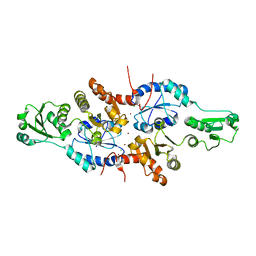 | | human ERp44 zinc-bound form | | 分子名称: | CHLORIDE ION, Endoplasmic reticulum resident protein 44, ZINC ION | | 著者 | Watanabe, S, Harayama, M, Inaba, K. | | 登録日 | 2017-06-30 | | 公開日 | 2019-01-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Zinc regulates ERp44-dependent protein quality control in the early secretory pathway.
Nat Commun, 10, 2019
|
|
4Z9V
 
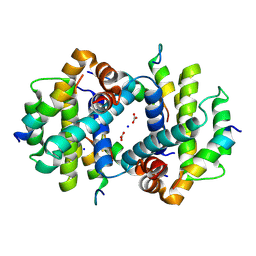 | | TCTP contains a BH3-like domain, which instead of inhibiting, activates Bcl-xL | | 分子名称: | BICARBONATE ION, Bcl-2-like protein 1,APOPTOSIS REGULATOR BCL-XL, SODIUM ION, ... | | 著者 | Cura, V, Agez, M, Thebault, S, Cavarelli, J. | | 登録日 | 2015-04-12 | | 公開日 | 2016-02-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.099 Å) | | 主引用文献 | TCTP contains a BH3-like domain, which instead of inhibiting, activates Bcl-xL.
Sci Rep, 6, 2016
|
|
2YWR
 
 | | Crystal structure of GAR transformylase from Aquifex aeolicus | | 分子名称: | COBALT (II) ION, MAGNESIUM ION, Phosphoribosylglycinamide formyltransferase | | 著者 | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-04-23 | | 公開日 | 2007-10-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Structures and reaction mechanisms of the two related enzymes, PurN and PurU.
J.Biochem., 154, 2013
|
|
