7P81
 
 | | Crystal structure of ClpP from Bacillus subtilis in complex with ADEP2 (compact state) | | Descriptor: | ADEP2, ATP-dependent Clp protease proteolytic subunit | | Authors: | Lee, B.-G, Kim, L, Kim, M.K, Kwon, D.H, Song, H.K. | | Deposit date: | 2021-07-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
1DGS
 
 | | CRYSTAL STRUCTURE OF NAD+-DEPENDENT DNA LIGASE FROM T. FILIFORMIS | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA LIGASE, ZINC ION | | Authors: | Lee, J.Y, Chang, C, Song, H.K, Kwon, S.T, Suh, S.W. | | Deposit date: | 1999-11-25 | | Release date: | 2000-11-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications.
EMBO J., 19, 2000
|
|
1V9P
 
 | | Crystal Structure Of Nad+-Dependent DNA Ligase | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA ligase, ZINC ION | | Authors: | Lee, J.Y, Chang, C, Song, H.K, Moon, J, Yang, J.K, Kim, H.K, Kwon, S.K, Suh, S.W. | | Deposit date: | 2004-01-27 | | Release date: | 2004-03-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications.
Embo J., 19, 2000
|
|
4XSJ
 
 | | Crystal structure of the N-terminal domain of the human mitochondrial calcium uniporter fused with T4 lysozyme | | Descriptor: | Lysozyme,Calcium uniporter protein, mitochondrial, SULFATE ION | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-01-22 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
4XTB
 
 | | Crystal structure of the N-terminal domain of the human mitochondrial calcium uniporter | | Descriptor: | Calcium uniporter protein, mitochondrial, TETRAETHYLENE GLYCOL | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-01-23 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
1AUR
 
 | |
1AUO
 
 | | CARBOXYLESTERASE FROM PSEUDOMONAS FLUORESCENS | | Descriptor: | CARBOXYLESTERASE | | Authors: | Kim, K.K, Song, H.K, Suh, S.W. | | Deposit date: | 1997-09-01 | | Release date: | 1998-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of carboxylesterase from Pseudomonas fluorescens, an alpha/beta hydrolase with broad substrate specificity.
Structure, 5, 1997
|
|
1DO0
 
 | | ORTHORHOMBIC CRYSTAL FORM OF HEAT SHOCK LOCUS U (HSLU) FROM ESCHERICHIA COLI | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PROTEIN (HEAT SHOCK LOCUS U), ... | | Authors: | Bochtler, M, Hartmann, C, Song, H.K, Bourenkov, G.P, Bartunik, H.D. | | Deposit date: | 1999-12-18 | | Release date: | 2000-02-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structures of HsIU and the ATP-dependent protease HsIU-HsIV.
Nature, 403, 2000
|
|
1JXV
 
 | | Crystal Structure of Human Nucleoside Diphosphate Kinase A | | Descriptor: | Nucleoside Diphosphate Kinase A | | Authors: | Min, K, Song, H.K, Chang, C, Kim, S.Y, Lee, K.J, Suh, S.W. | | Deposit date: | 2001-09-10 | | Release date: | 2002-04-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human nucleoside diphosphate kinase A, a metastasis suppressor.
Proteins, 46, 2002
|
|
4EBR
 
 | | Crystal structure of Autophagic E2, Atg10 | | Descriptor: | MERCURY (II) ION, Ubiquitin-like-conjugating enzyme ATG10 | | Authors: | Hong, S.B, Kim, B.W, Kim, J.H, Song, H.K. | | Deposit date: | 2012-03-24 | | Release date: | 2012-10-03 | | Last modified: | 2013-07-24 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Structure of the autophagic E2 enzyme Atg10
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4FBA
 
 | | Structure of mutant RIP from barley seeds in complex with adenine | | Descriptor: | ADENINE, Protein synthesis inhibitor I | | Authors: | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | Deposit date: | 2012-05-22 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4FBB
 
 | | Structure of mutant RIP from barley seeds in complex with adenine (AMP-incubated) | | Descriptor: | ADENINE, Protein synthesis inhibitor I | | Authors: | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | Deposit date: | 2012-05-22 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4FBH
 
 | | Structure of RIP from barley seeds | | Descriptor: | ADENOSINE MONOPHOSPHATE, Protein synthesis inhibitor I | | Authors: | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4FBC
 
 | | Structure of mutant RIP from barley seeds in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Protein synthesis inhibitor I | | Authors: | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | Deposit date: | 2012-05-22 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4FB9
 
 | | Structure of mutant RIP from barley seeds | | Descriptor: | Protein synthesis inhibitor I | | Authors: | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | Deposit date: | 2012-05-22 | | Release date: | 2012-10-31 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4GQW
 
 | |
4HAN
 
 | | Crystal structure of Galectin 8 with NDP52 peptide | | Descriptor: | Calcium-binding and coiled-coil domain-containing protein 2, DI(HYDROXYETHYL)ETHER, Galectin-8, ... | | Authors: | Kim, B.-W, Hong, S.B, Kim, J.H, Kwon, D.H, Song, H.K. | | Deposit date: | 2012-09-27 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Structural basis for recognition of autophagic receptor NDP52 by the sugar receptor galectin-8.
Nat Commun, 4, 2013
|
|
4GQV
 
 | | Crystal structure of CBS-pair protein, CBSX1 from Arabidopsis thaliana | | Descriptor: | CBS domain-containing protein CBSX1, chloroplastic | | Authors: | Jeong, B.-C, Park, S.H, Yoo, K.S, Shin, J.S, Song, H.K. | | Deposit date: | 2012-08-24 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Crystal structure of the single cystathionine beta-synthase domain-containing protein CBSX1 from Arabidopsis thaliana
Biochem.Biophys.Res.Commun., 430, 2013
|
|
1EW4
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI CYAY PROTEIN REVEALS A NOVEL FOLD FOR THE FRATAXIN FAMILY | | Descriptor: | CYAY PROTEIN | | Authors: | Suh, S.W, Cho, S, Lee, M.G, Yang, J.K, Lee, J.Y, Song, H.K. | | Deposit date: | 2000-04-22 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of Escherichia coli CyaY protein reveals a previously unidentified fold for the evolutionarily conserved frataxin family.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
5K5U
 
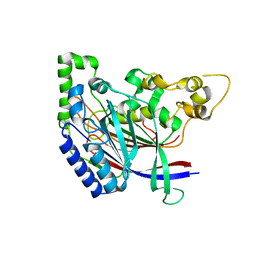 | |
5K63
 
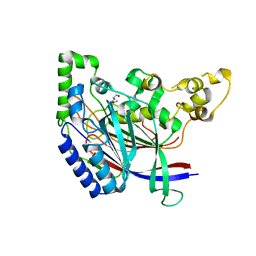 | | Crystal structure of N-terminal amidase C187S | | Descriptor: | ASPARAGINE, GLYCINE, Nta1p | | Authors: | Kim, M.K, Oh, S.-J, Lee, B.-G, Song, H.K. | | Deposit date: | 2016-05-24 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for dual specificity of yeast N-terminal amidase in the N-end rule pathway.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5JST
 
 | | MBP fused MDV1 coiled coil | | Descriptor: | ACETATE ION, GLYCEROL, Maltose-binding periplasmic protein,Mitochondrial division protein 1, ... | | Authors: | Kim, B.-W, Song, H.K. | | Deposit date: | 2016-05-09 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | ACCORD: an assessment tool to determine the orientation of homodimeric coiled-coils.
Sci Rep, 7, 2017
|
|
1FK2
 
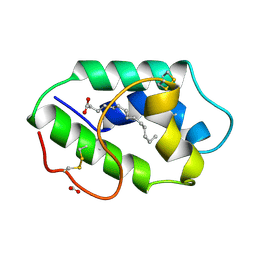 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH MYRISTIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, MYRISTIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK7
 
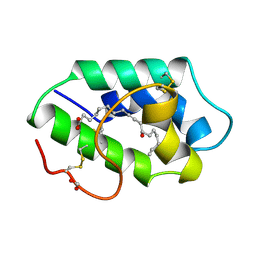 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH RICINOLEIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, NON-SPECIFIC LIPID TRANSFER PROTEIN, RICINOLEIC ACID | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK3
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH PALMITOLEIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN, PALMITOLEIC ACID | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
