6RLS
 
 | | Concerted dynamics of metallo-base pairs in an A/B-form helical transition (apo species) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*TP*CP*AP*TP*GP*AP*TP*AP*CP*G)-3')_apo | | Authors: | Schmidt, O.P, Jurt, S, Johannsen, S, Karimi, A, Sigel, R.K.O, Luedtke, N.W. | | Deposit date: | 2019-05-02 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Concerted dynamics of metallo-base pairs in an A/B-form helical transition.
Nat Commun, 10, 2019
|
|
2AHT
 
 | |
6EZ0
 
 | | Specific phosphorothioate substitution within domain 6 of a group II intron ribozyme leads to changes in local structure and metal ion binding | | Descriptor: | RNA (27-MER) | | Authors: | Erat, M.C, Besic, E, Oberhuber, M, Johannsen, S, Sigel, R.K.O. | | Deposit date: | 2017-11-13 | | Release date: | 2018-01-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Specific phosphorothioate substitution within domain 6 of a group II intron ribozyme leads to changes in local structure and metal ion binding.
J. Biol. Inorg. Chem., 23, 2018
|
|
6GN4
 
 | | tc-DNA/tc-DNA duplex | | Descriptor: | Tc-DNA (5'-D(*(TCJ)P*(TTK)P*(TCJ)P*(TCS)P*(TCS)P*(TCJ)P*(TTK)P*(TTK)P*(TCY)P*(TCJ))-3'), Tc-DNA (5'-D(*(TCS)P*(TTK)P*(TCY)P*(TCY)P*(TCS)P*(TCJ)P*(TCJ)P*(TCS)P*(TCY)P*(TCS))-3') | | Authors: | Istrate, A, Johannsen, S, Istrate, A, Sigel, R.K.O, Leumann, C. | | Deposit date: | 2018-05-29 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of tricyclo-DNA containing duplexes: insight into enhanced thermal stability and nuclease resistance.
Nucleic Acids Res., 47, 2019
|
|
6GMY
 
 | | Tc-DNA/RNA duplex | | Descriptor: | RNA (5'-R(*GP*UP*AP*AP*GP*CP*CP*GP*AP*G)-3'), Tc-DNA (5'-(*(TCJ)P*(TTK)P*(TCJ)P*(TCS)P*(TCS)P*(TCJ)P*(TTK)P*(TTK)P*(TCY)P*(TCJ))-3') | | Authors: | Istrate, A, Johannsen, S, Istrate, A, Sigel, R.K.O, Leumann, C. | | Deposit date: | 2018-05-28 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of tricyclo-DNA containing duplexes: insight into enhanced thermal stability and nuclease resistance.
Nucleic Acids Res., 47, 2019
|
|
6GPI
 
 | | Tc-DNA/DNA duplex | | Descriptor: | DNA (5'-D(*GP*TP*AP*AP*GP*CP*CP*GP*AP*G)-3'), tc-DNA (5'-D(*(TCJ)P*(TTK)P*(TCJ)P*(TCS)P*(TCS)P*(TCJ)P*(TTK)P*(TTK)P*(TCY)P*(TCJ))-3') | | Authors: | Istrate, A, Johannsen, S, Istrate, A, Sigel, R.K.O, Leumann, C. | | Deposit date: | 2018-06-05 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of tricyclo-DNA containing duplexes: insight into enhanced thermal stability and nuclease resistance.
Nucleic Acids Res., 47, 2019
|
|
6FY6
 
 | | Concerted dynamics of metallo-base pairs in an A/B-form helical transition (major species) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*TP*CP*AP*TP*GP*AP*TP*AP*CP*G)-3')_major, MERCURY (II) ION | | Authors: | Schmidt, O.P, Jurt, S, Johannsen, S, Karimi, A, Sigel, R.K.O, Luedtke, N.W. | | Deposit date: | 2018-03-11 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Concerted dynamics of metallo-base pairs in an A/B-form helical transition.
Nat Commun, 10, 2019
|
|
6FY7
 
 | | Concerted dynamics of metallo-base pairs in an A/B-form helical transition (minor species) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*TP*CP*AP*TP*GP*AP*TP*AP*CP*G)-3')_minor, MERCURY (II) ION | | Authors: | Schmidt, O.P, Jurt, S, Johannsen, S, Karimi, A, Sigel, R.K.O, Luedtke, N.W. | | Deposit date: | 2018-03-11 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Concerted dynamics of metallo-base pairs in an A/B-form helical transition.
Nat Commun, 10, 2019
|
|
7Q48
 
 | |
7QA2
 
 | |
7Q6L
 
 | |
2K69
 
 | |
2K68
 
 | |
2K65
 
 | |
2K67
 
 | |
2LI4
 
 | |
2LU0
 
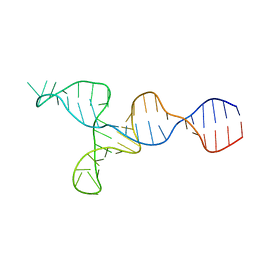 | | NMR solution structure of the kappa-zeta region of S.cerevisiae group II intron ai5(gamma) | | Descriptor: | RNA (49-MER) | | Authors: | Donghi, D, Pechlaner, M, Finazzo, C, Knobloch, B, Sigel, R.K.O. | | Deposit date: | 2012-06-05 | | Release date: | 2013-04-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structural stabilization of the kappa three-way junction by Mg(II) represents the first step in the folding of a group II intron.
Nucleic Acids Res., 41, 2013
|
|
2M57
 
 | |
1R2P
 
 | | Solution structure of domain 5 from the ai5(gamma) group II intron | | Descriptor: | 34-MER | | Authors: | Sigel, R.K.O, Sashital, D.G, Abramovitz, D.L, Palmer III, A.G, Butcher, S.E, Pyle, A.M. | | Deposit date: | 2003-09-29 | | Release date: | 2004-02-03 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of domain 5 of a group II intron ribozyme reveals a new RNA motif.
Nat.Struct.Mol.Biol., 11, 2004
|
|
4JIY
 
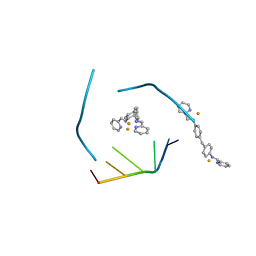 | | RNA three-way junction stabilized by a supramolecular di-iron(II) cylinder drug | | Descriptor: | 5'-(CGUACG)-3', FE (II) ION, N-[(1E)-PYRIDIN-2-YLMETHYLENE]-N-[4-(4-{[(1E)-PYRIDIN-2-YLMETHYLENE]AMINO}BENZYL)PHENYL]AMINE | | Authors: | Sigel, R.K.O, Schnabl, J.A, Freisinger, E, Spingler, B, Hannon, M.J. | | Deposit date: | 2013-03-07 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Binding of a designed anti-cancer drug to the central cavity of an RNA three-way junction.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4U78
 
 | | Octameric RNA duplex soaked in copper(II)chloride | | Descriptor: | CALCIUM ION, COPPER (II) ION, RNA (5'-R(*UP*CP*GP*UP*AP*CP*GP*A)-3') | | Authors: | Schaffer, M.F, Spingler, B, Schnabl, J, Peng, G, Olieric, V, Sigel, R.K.O. | | Deposit date: | 2014-07-30 | | Release date: | 2015-08-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | The X-ray Structures of Six Octameric RNA Duplexes in the Presence of Different Di- and Trivalent Cations.
Int J Mol Sci, 17, 2016
|
|
4U47
 
 | | Octameric RNA duplex soaked in terbium(III)chloride | | Descriptor: | RNA (5'-R(*UP*CP*GP*UP*AP*CP*GP*A)-3'), TERBIUM(III) ION | | Authors: | Schaffer, M.F, Spingler, B, Schnabl, J, Peng, G, Olieric, V, Sigel, R.K.O. | | Deposit date: | 2014-07-23 | | Release date: | 2015-08-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | The X-ray Structures of Six Octameric RNA Duplexes in the Presence of Different Di- and Trivalent Cations.
Int J Mol Sci, 17, 2016
|
|
4U3P
 
 | | Octameric RNA duplex co-crystallized with strontium(II)chloride | | Descriptor: | RNA (5'-R(*UP*CP*GP*UP*AP*CP*GP*A)-3'), STRONTIUM ION | | Authors: | Schaffer, M.F, Spingler, B, Schnabl, J, Peng, G, Olieric, V, Sigel, R.K.O. | | Deposit date: | 2014-07-22 | | Release date: | 2015-07-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.866 Å) | | Cite: | The X-ray Structures of Six Octameric RNA Duplexes in the Presence of Different Di- and Trivalent Cations.
Int J Mol Sci, 17, 2016
|
|
4U3R
 
 | | Octameric RNA duplex co-crystallized with cobalt(II)chloride | | Descriptor: | COBALT (II) ION, RNA (5'-R(*UP*CP*GP*UP*AP*CP*GP*A)-3') | | Authors: | Schaffer, M.F, Spingler, B, Schnabl, J, Peng, G, Olieric, V, Sigel, R.K.O. | | Deposit date: | 2014-07-22 | | Release date: | 2015-07-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | The X-ray Structures of Six Octameric RNA Duplexes in the Presence of Different Di- and Trivalent Cations.
Int J Mol Sci, 17, 2016
|
|
4U3L
 
 | | octameric RNA duplex co-crystallized in calcium(II)chloride | | Descriptor: | CALCIUM ION, RNA (5'-R(*UP*CP*GP*UP*AP*CP*GP*A)-3') | | Authors: | Schaffer, M.F, Spingler, B, Schnabl, J, Peng, G, Olieric, V, Sigel, R.K.O. | | Deposit date: | 2014-07-22 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | The X-ray Structures of Six Octameric RNA Duplexes in the Presence of Different Di- and Trivalent Cations.
Int J Mol Sci, 17, 2016
|
|
