2ENU
 
 | |
2EMR
 
 | |
2EN5
 
 | |
2E4N
 
 | |
2EH4
 
 | |
2ED5
 
 | |
2EH2
 
 | |
2ELE
 
 | |
2EH5
 
 | |
2ZVB
 
 | |
2ZV3
 
 | |
2ZVC
 
 | |
2RQZ
 
 | | Structure of sugar modified epidermal growth factor-like repeat 12 of mouse Notch-1 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-alpha-L-fucopyranose, Neurogenic locus notch homolog protein 1 | | Authors: | Shimizu, K, Fujitani, N, Hosoguchi, K, Nishimura, S. | | Deposit date: | 2010-02-26 | | Release date: | 2010-10-13 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Chemical Synthesis, Folding, and Structural Insights into O-Fucosylated Epidermal Growth Factor-like Repeat 12 of Mouse Notch-1 Receptor
J.Am.Chem.Soc., 132, 2010
|
|
2RR2
 
 | | Structure of O-fucosylated epidermal growth factor-like repeat 12 of mouse Notch-1 receptor | | Descriptor: | Neurogenic locus notch homolog protein 1, alpha-L-fucopyranose | | Authors: | Hosoguchi, K, Shimizu, K, Fujitani, N, Nishimura, S. | | Deposit date: | 2010-02-26 | | Release date: | 2010-10-13 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Chemical Synthesis, Folding, and Structural Insights into O-Fucosylated Epidermal Growth Factor-like Repeat 12 of Mouse Notch-1 Receptor
J.Am.Chem.Soc., 132, 2010
|
|
2RR0
 
 | | Structure of epidermal growth factor-like repeat 12 of mouse Notch-1 receptor | | Descriptor: | Neurogenic locus notch homolog protein 1 | | Authors: | Hosoguchi, K, Shimizu, K, Fujitani, N, Nishimura, S. | | Deposit date: | 2010-02-26 | | Release date: | 2010-10-13 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Chemical Synthesis, Folding, and Structural Insights into O-Fucosylated Epidermal Growth Factor-like Repeat 12 of Mouse Notch-1 Receptor
J.Am.Chem.Soc., 132, 2010
|
|
5B2C
 
 | | Crystal structure of Mumps virus hemagglutinin-neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HN protein, SULFATE ION | | Authors: | Kubota, M, Takeuchi, K, Watanabe, S, Ohno, S, Matsuoka, R, Kohda, D, Hiramatsu, H, Suzuki, Y, Nakayama, T, Terada, T, Shimizu, K, Shimizu, N, Yanagi, Y, Hashiguchi, T. | | Deposit date: | 2016-01-14 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.238 Å) | | Cite: | Trisaccharide containing alpha 2,3-linked sialic acid is a receptor for mumps virus
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5B2D
 
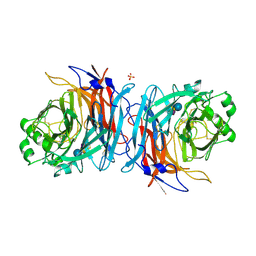 | | Crystal structure of Mumps virus hemagglutinin-neuraminidase bound to 3-sialyllactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HN protein, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Kubota, M, Takeuchi, K, Watanabe, S, Ohno, S, Matsuoka, R, Kohda, D, Hiramatsu, H, Suzuki, Y, Nakayama, T, Terada, T, Shimizu, K, Shimizu, N, Yanagi, Y, Hashiguchi, T. | | Deposit date: | 2016-01-14 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | Trisaccharide containing alpha 2,3-linked sialic acid is a receptor for mumps virus
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6IGX
 
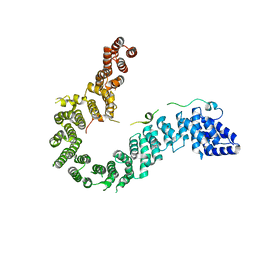 | | Crystal structure of human CAP-G in complex with CAP-H | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Condensin complex subunit 2, Condensin complex subunit 3 | | Authors: | Hara, K, Migita, T, Shimizu, K, Hashimoto, H. | | Deposit date: | 2018-09-26 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.995 Å) | | Cite: | Structural basis of HEAT-kleisin interactions in the human condensin I subcomplex.
Embo Rep., 20, 2019
|
|
7FBO
 
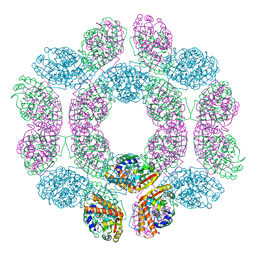 | | geranyl pyrophosphate C6-methyltransferase BezA binding with S-adenosylhomocysteine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BezA, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Tsutsumi, H, Moriwaki, Y, Terada, T, Shimizu, K, Katsuyama, Y, Ohnishi, Y. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural and Molecular Basis of the Catalytic Mechanism of Geranyl Pyrophosphate C6-Methyltransferase: Creation of an Unprecedented Farnesyl Pyrophosphate C6-Methyltransferase.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7FBH
 
 | | geranyl pyrophosphate C6-methyltransferase BezA | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BezA, ... | | Authors: | Tsutsumi, H, Moriwaki, Y, Terada, T, Shimizu, K, Katsuyama, Y, Ohnishi, Y. | | Deposit date: | 2021-07-10 | | Release date: | 2021-12-01 | | Last modified: | 2022-03-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Molecular Basis of the Catalytic Mechanism of Geranyl Pyrophosphate C6-Methyltransferase: Creation of an Unprecedented Farnesyl Pyrophosphate C6-Methyltransferase.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
1WNG
 
 | |
1WQL
 
 | | Cumene dioxygenase (cumA1A2) from Pseudomonas fluorescens IP01 | | Descriptor: | FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, OXYGEN MOLECULE, ... | | Authors: | Dong, X, Fushinobu, S, Fukuda, E, Terada, T, Nakamura, S, Shimizu, K, Nojiri, H, Omori, T, Shoun, H, Wakagi, T. | | Deposit date: | 2004-09-30 | | Release date: | 2005-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Terminal Oxygenase Component of Cumene Dioxygenase from Pseudomonas fluorescens IP01
J.BACTERIOL., 187, 2005
|
|
1YT6
 
 | | NMR structure of peptide SD | | Descriptor: | peptide SD | | Authors: | Murata, T, Hemmi, H, Nakamura, S, Shimizu, K, Suzuki, Y, Yamaguchi, I. | | Deposit date: | 2005-02-10 | | Release date: | 2005-09-27 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure, epitope mapping, and docking simulation of a gibberellin mimic peptide as a peptidyl mimotope for a hydrophobic ligand.
Febs J., 272, 2005
|
|
1VCE
 
 | |
2DCT
 
 | | Crystal structure of the TT1209 from Thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, SODIUM ION, hypothetical protein TTHA0104 | | Authors: | Asada, Y, Sugahara, M, Shimizu, K, Yamamoto, H, Shimada, H, Nakamoto, T, Ono, N, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-01-12 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of the TT1209 from Thermus thermophilus HB8
To be Published
|
|
