8H97
 
 | | GH86 agarase Aga86A_Wa | | Descriptor: | Beta-agarase, CALCIUM ION, HEXAETHYLENE GLYCOL | | Authors: | Zhang, Y.Y, Dong, S, Feng, Y.G, Chang, Y.G. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural characterization on a beta-agarase Aga86A_Wa from Wenyingzhuangia aestuarii reveals the prevalent methyl-galactose accommodation capacity of GH86 enzymes at subsite -1.
Carbohydr Polym, 306, 2023
|
|
7DPV
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with 7-O-methyl-dihydromyricetin | | Descriptor: | (2S,3S)-3,5-dihydroxy-7-methoxy-2-(3,4,5-trihydroxyphenyl)chroman-4-one, 3C-like proteinase | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease.
Nat Commun, 12, 2021
|
|
7DPP
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with myricetin | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, 3C-like proteinase | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease.
Nat Commun, 12, 2021
|
|
7DPU
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with 7-O-methyl-myricetin | | Descriptor: | 3C-like proteinase, 7-methoxy-3,5-bis(oxidanyl)-2-[3,4,5-tris(oxidanyl)phenyl]chromen-4-one, GLYCEROL | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease.
Nat Commun, 12, 2021
|
|
6KIK
 
 | | Crystal structure of a thermostable aldo-keto reductase Tm1743 in complex with inhibitor tolrestat | | Descriptor: | Oxidoreductase, aldo/keto reductase family, TOLRESTAT | | Authors: | Zhang, C.Y, Liu, X.M, Wang, C, Tang, W.R. | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Tolrestat acts atypically as a competitive inhibitor of the thermostable aldo-keto reductase Tm1743 from Thermotoga maritima.
Febs Lett., 594, 2020
|
|
6KIY
 
 | | Crystal structure of a thermostable aldo-keto reductase Tm1743 in complex with inhibitor Epalrestat | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase, aldo/keto reductase family, ... | | Authors: | Zhang, C.Y, Liu, X.M, Wang, C, Min, Z.Z, Xu, X.L. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tolrestat acts atypically as a competitive inhibitor of the thermostable aldo-keto reductase Tm1743 from Thermotoga maritima.
Febs Lett., 594, 2020
|
|
6KY6
 
 | | Crystal structure of a thermostable aldo-keto reductase Tm1743 in complexs with inhibitor epalrestat in space group P3221cc | | Descriptor: | 2,5-diketo-D-gluconic acid reductase, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zhang, C.Y, Min, Z.Z, Liu, X.M, Wang, C, Tang, W.R. | | Deposit date: | 2019-09-16 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Tolrestat acts atypically as a competitive inhibitor of the thermostable aldo-keto reductase Tm1743 from Thermotoga maritima.
Febs Lett., 594, 2020
|
|
8IBQ
 
 | | Bromodomain and Extra-terminal Domain (BET) BRD4 | | Descriptor: | 7-[2-fluoranyl-3-(1,3,5-trimethylpyrazol-4-yl)phenyl]-1~{H}-imidazo[4,5-b]pyridine, Bromodomain-containing protein 4 | | Authors: | Cao, D, Zhiyan, D, Xiong, B. | | Deposit date: | 2023-02-10 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of 1 H -Imidazo[4,5- b ]pyridine Derivatives as Potent and Selective BET Inhibitors for the Management of Neuropathic Pain.
J.Med.Chem., 66, 2023
|
|
8IDH
 
 | | Bromodomain and Extra-terminal Domain (BET) BRD4 | | Descriptor: | 7-[2-fluoranyl-3-(1,3,5-trimethylpyrazol-4-yl)phenyl]-1~{H}-imidazo[4,5-b]pyridine, Bromodomain-containing protein 4 | | Authors: | Cao, D, Zhiyan, D, Xiong, B. | | Deposit date: | 2023-02-13 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Discovery of 1 H -Imidazo[4,5- b ]pyridine Derivatives as Potent and Selective BET Inhibitors for the Management of Neuropathic Pain.
J.Med.Chem., 66, 2023
|
|
8IGX
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 9 (simnotrelvir, SIM0417, SSD8432) | | Descriptor: | (8~{S})-~{N}-[(1~{S})-1-cyano-2-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]ethyl]-7-[(2~{S})-3,3-dimethyl-2-[2,2,2-tris(fluoranyl)ethanoylamino]butanoyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFS
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 7 | | Descriptor: | (8~{S})-7-[(2~{S})-2-(~{tert}-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-~{N}-[(1~{S})-1-cyano-2-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]ethyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFP
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 1 | | Descriptor: | (1R,2S,5S)-3-[(2S)-2-(tert-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-6,6-dimethyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFQ
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 2 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFR
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 3 | | Descriptor: | (1R,2S,5S)-3-[(2S)-2-(tert-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-6,6-dimethyl-N-[(2S)-5-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]hex-3-en-2-yl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFT
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 10 | | Descriptor: | (8S)-N-[(1S)-1-cyano-2-[(3S)-2-oxidanylidenepyrrolidin-3-yl]ethyl]-7-[(2S)-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IGY
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
6ML6
 
 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 4 with a CpA 5mC Modification) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*(5CM)P*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*CP*GP*AP*AP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*TP*TP*CP*GP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
6ML5
 
 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 4) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*CP*GP*AP*AP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*TP*TP*CP*GP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
6ML4
 
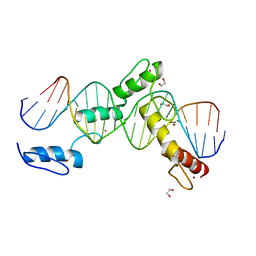 | | BTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 3) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*CP*GP*AP*AP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*GP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
6ML2
 
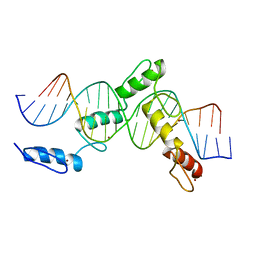 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 1) | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*CP*AP*GP*CP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ZINC ION, ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.874 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
6ML7
 
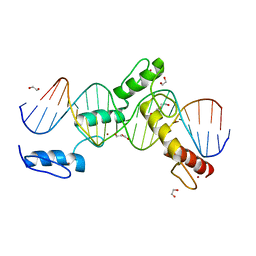 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 4 with a CpG 5mC Modification) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*(5CM)P*GP*AP*AP*TP*T)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
6ML3
 
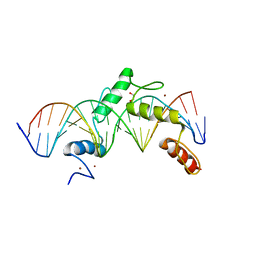 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 2) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*AP*GP*CP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*GP*CP*TP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.683 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
7YZY
 
 | | pMMO structure from native membranes by cryoET and STA | | Descriptor: | Methane monooxygenase subunit C2, Particulate methane monooxygenase alpha subunit, Particulate methane monooxygenase beta subunit | | Authors: | Zhu, Y, Ni, T, Zhang, P. | | Deposit date: | 2022-02-21 | | Release date: | 2022-08-10 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure and activity of particulate methane monooxygenase arrays in methanotrophs.
Nat Commun, 13, 2022
|
|
