7TMT
 
 | | V-ATPase from Saccharomyces cerevisiae, State 3 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, H(+)-transporting two-sector ATPase, V-ATPase subunit E, ... | | 著者 | Vasanthakumar, T, Keon, K.A, Bueler, S.A, Jaskolka, M.C, Rubinstein, J.L. | | 登録日 | 2022-01-20 | | 公開日 | 2022-04-20 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Coordinated conformational changes in the V 1 complex during V-ATPase reversible dissociation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5TJ5
 
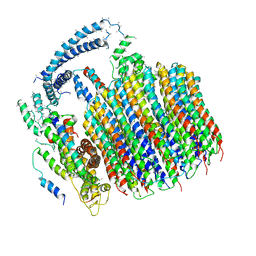 | | Atomic model for the membrane-embedded motor of a eukaryotic V-ATPase | | 分子名称: | V-type proton ATPase subunit a, V-type proton ATPase subunit c, V-type proton ATPase subunit c', ... | | 著者 | Mazhab-Jafari, M.T, Rohou, A, Schmidt, C, Bueler, S.A, Benlekbir, S, Robinson, C.V, Rubinstein, J.L. | | 登録日 | 2016-10-03 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Atomic model for the membrane-embedded VO motor of a eukaryotic V-ATPase.
Nature, 539, 2016
|
|
5VCA
 
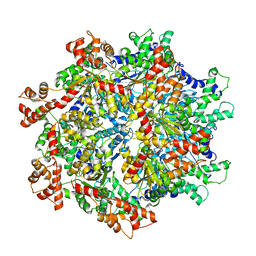 | | VCP like ATPase from T. acidophilum (VAT)-Substrate bound conformation | | 分子名称: | VCP-like ATPase | | 著者 | Ripstein, Z.A, Huang, R, Augustyniak, R, Kay, L.E, Rubinstein, J.L. | | 登録日 | 2017-03-31 | | 公開日 | 2017-04-26 | | 最終更新日 | 2020-01-15 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Structure of a AAA+ unfoldase in the process of unfolding substrate.
Elife, 6, 2017
|
|
5VC7
 
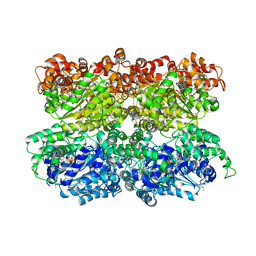 | | VCP like ATPase from T. acidophilum (VAT) - conformation 1 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, VCP-like ATPase | | 著者 | Ripstein, Z.A, Huang, R, Augustyniak, R, Kay, L.E, Rubinstein, J.L. | | 登録日 | 2017-03-31 | | 公開日 | 2017-04-26 | | 最終更新日 | 2020-01-15 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structure of a AAA+ unfoldase in the process of unfolding substrate.
Elife, 6, 2017
|
|
7RJE
 
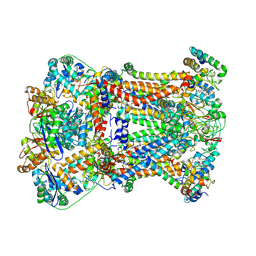 | | Complex III2 from Candida albicans, Inz-5 bound | | 分子名称: | 3-[2-fluoro-5-(trifluoromethyl)phenyl]-7-methyl-1-[(2-methyl-2H-tetrazol-5-yl)methyl]-1H-indazole, Cytochrome b, Cytochrome b-c1 complex subunit 2, ... | | 著者 | Di Trani, J.M, Rubinstein, J.L. | | 登録日 | 2021-07-20 | | 公開日 | 2021-09-15 | | 最終更新日 | 2022-01-19 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Rieske head domain dynamics and indazole-derivative inhibition of Candida albicans complex III.
Structure, 30, 2022
|
|
7RJC
 
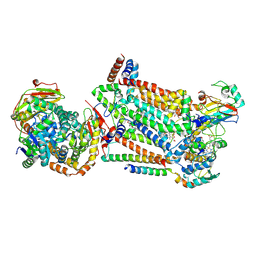 | | Complex III2 from Candida albicans, inhibitor free, Rieske head domain in intermediate position | | 分子名称: | Cytochrome b, Cytochrome b-c1 complex subunit 2, mitochondrial, ... | | 著者 | Di Trani, J.M, Rubinstein, J.L. | | 登録日 | 2021-07-20 | | 公開日 | 2021-09-15 | | 最終更新日 | 2022-01-19 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Rieske head domain dynamics and indazole-derivative inhibition of Candida albicans complex III.
Structure, 30, 2022
|
|
7RJD
 
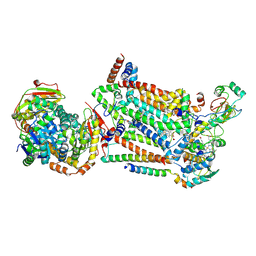 | | Complex III2 from Candida albicans, inhibitor free, Rieske head domain in c position | | 分子名称: | Cytochrome b, Cytochrome b-c1 complex subunit 2, mitochondrial, ... | | 著者 | Di Trani, J.M, Rubinstein, J.L. | | 登録日 | 2021-07-20 | | 公開日 | 2021-09-15 | | 最終更新日 | 2022-01-19 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Rieske head domain dynamics and indazole-derivative inhibition of Candida albicans complex III.
Structure, 30, 2022
|
|
7RJB
 
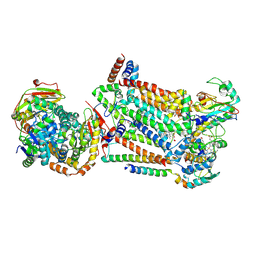 | | Complex III2 from Candida albicans, inhibitor free, Rieske head domain in b position | | 分子名称: | Cytochrome b, Cytochrome b-c1 complex subunit 2, mitochondrial, ... | | 著者 | Di Trani, J.M, Rubinstein, J.L. | | 登録日 | 2021-07-20 | | 公開日 | 2021-09-15 | | 最終更新日 | 2022-01-19 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Rieske head domain dynamics and indazole-derivative inhibition of Candida albicans complex III.
Structure, 30, 2022
|
|
7RJA
 
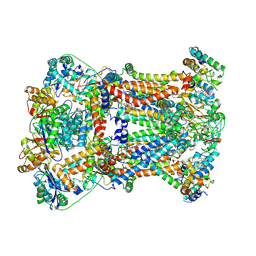 | |
6OLJ
 
 | |
4EOZ
 
 | |
8U7Z
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: Local Refinment of KCTD5(CTD)/Gbeta1gamma2 | | 分子名称: | BTB/POZ domain-containing protein KCTD5, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | | 著者 | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | 登録日 | 2023-09-15 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.97 Å) | | 主引用文献 | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U80
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: Local Refinment of KCTD5(BTB)/Cullin3(NTD) | | 分子名称: | BTB/POZ domain-containing protein KCTD5, Cullin-3 | | 著者 | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | 登録日 | 2023-09-15 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U83
 
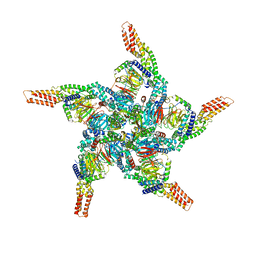 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State C From Composite RELION Multi-body Refinement Map | | 分子名称: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | 登録日 | 2023-09-15 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.975 Å) | | 主引用文献 | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U81
 
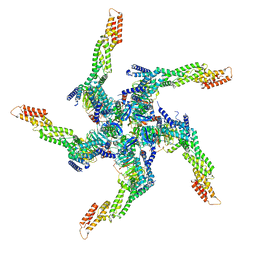 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State A From Composite RELION Multi-body Refinement Map | | 分子名称: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | 登録日 | 2023-09-15 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.82 Å) | | 主引用文献 | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U82
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State B From Composite RELION Multi-body Refinement Map | | 分子名称: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | 登録日 | 2023-09-15 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.84 Å) | | 主引用文献 | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U84
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State D From Composite RELION Multi-body Refinement Map | | 分子名称: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | 登録日 | 2023-09-15 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.88 Å) | | 主引用文献 | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6X87
 
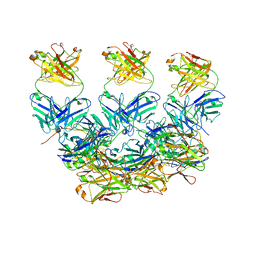 | | CryoEM structure of the Plasmodium berghei circumsporozoite protein in complex with inhibitory mouse antibody 3D11. | | 分子名称: | 3D11 Fab heavy chain, 3D11 Fab kappa chain, Circumsporozoite protein | | 著者 | Kucharska, I, Thai, E, Rubinstein, J, Julien, J.P. | | 登録日 | 2020-06-01 | | 公開日 | 2020-12-02 | | 最終更新日 | 2020-12-16 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural ordering of the Plasmodium berghei circumsporozoite protein repeats by inhibitory antibody 3D11.
Elife, 9, 2020
|
|
8D4T
 
 | | Mammalian CIV with GDN bound | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, ... | | 著者 | Di Trani, J, Rubinstein, J. | | 登録日 | 2022-06-02 | | 公開日 | 2022-07-06 | | 最終更新日 | 2022-08-24 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of mammalian complex IV inhibition by steroids.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3B8N
 
 | |
3B8O
 
 | |
3B8M
 
 | |
3B8P
 
 | |
2KX4
 
 | | Solution structure of Bacteriophage Lambda gpFII | | 分子名称: | Tail attachment protein | | 著者 | Maxwell, K.L, Cardarelli, L, Neudecker, P, Davidson, A.R, Ontario Centre for Structural Proteomics (OCSP) | | 登録日 | 2010-04-26 | | 公開日 | 2010-07-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Phages have adapted the same protein fold to fulfill multiple functions in virion assembly.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2LX4
 
 | |
