4Q9Q
 
 | | Crystal structure of an RNA aptamer bound to bromo-ligand analog in complex with Fab | | Descriptor: | (5Z)-5-(3-bromobenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, Fab BL3-6, HEAVY CHAIN, ... | | Authors: | Huang, H, Suslov, N.B, Li, N.-S, Shelke, S.A, Evans, M.E, Koldobskaya, Y, Rice, P.A, Piccirilli, J.A. | | Deposit date: | 2014-05-01 | | Release date: | 2014-06-18 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore.
Nat.Chem.Biol., 10, 2014
|
|
4Q9R
 
 | | Crystal structure of an RNA aptamer bound to trifluoroethyl-ligand analog in complex with Fab | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, Fab BL3-6, HEAVY CHAIN, ... | | Authors: | Huang, H, Suslov, N.B, Li, N.-S, Shelke, S.A, Evans, M.E, Koldobskaya, Y, Rice, P.A, Piccirilli, J.A. | | Deposit date: | 2014-05-01 | | Release date: | 2014-06-18 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore.
Nat.Chem.Biol., 10, 2014
|
|
5C7W
 
 | | 5'-monophosphate Z:P Guanine Riboswitch bound to hypoxanthine. | | Descriptor: | 5'-monophosphate Z:P guanine riboswitch, COBALT HEXAMMINE(III), HYPOXANTHINE | | Authors: | Hernandez, A.R, Shao, Y, Hoshika, S, Yang, Z, Shelke, S.A, Herrou, J, Kim, H.-J, Kim, M.-J, Piccirilli, J.A, Benner, S.A. | | Deposit date: | 2015-06-25 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | A Crystal Structure of a Functional RNA Molecule Containing an Artificial Nucleobase Pair.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5C7U
 
 | | 5'-monophosphate wt Guanine Riboswitch bound to hypoxanthine. | | Descriptor: | 5'-monophosphate wt guanine riboswitch, COBALT HEXAMMINE(III), HYPOXANTHINE | | Authors: | Hernandez, A.R, Shao, Y, Hoshika, S, Yang, Z, Shelke, S.A, Herrou, J, Kim, H.-J, Kim, M.-J, Piccirilli, J.A, Benner, S.A. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | A Crystal Structure of a Functional RNA Molecule Containing an Artificial Nucleobase Pair.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5E08
 
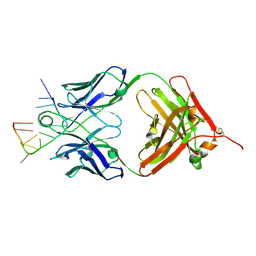 | | Specific Recognition of a Single-stranded RNA Sequence by an Engineered Synthetic Antibody Fragment | | Descriptor: | Fab Heavy Chain, Fab Light Chain, RNA | | Authors: | Huang, H, Qin, D, Li, N, Shao, Y, Staley, J.P, Kossiakoff, A.A, Koide, S, Piccirilli, J.A. | | Deposit date: | 2015-09-28 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Specific Recognition of a Single-Stranded RNA Sequence by a Synthetic Antibody Fragment.
J.Mol.Biol., 428, 2016
|
|
2KFZ
 
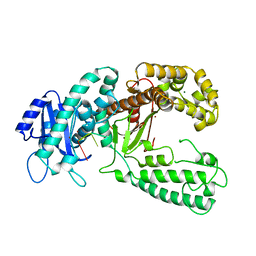 | | KLENOW FRAGMENT WITH BRIDGING-SULFUR SUBSTRATE AND ZINC ONLY | | Descriptor: | 5'-D(*GP*CP*TP*TP*AP*(US1)P*G)-3', KLENOW FRAGMENT OF DNA POLYMERASE I, MAGNESIUM ION, ... | | Authors: | Brautigam, C.A, Sun, S, Piccirilli, J.A, Steitz, T.A. | | Deposit date: | 1998-07-02 | | Release date: | 1998-11-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of normal single-stranded DNA and deoxyribo-3'-S-phosphorothiolates bound to the 3'-5' exonucleolytic active site of DNA polymerase I from Escherichia coli.
Biochemistry, 38, 1999
|
|
2KFN
 
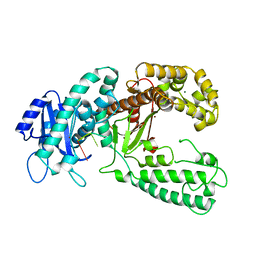 | | KLENOW FRAGMENT WITH BRIDGING-SULFUR SUBSTRATE AND MANGANESE | | Descriptor: | 5'-D(*GP*CP*TP*TP*AP*(US1)P*G)-3', KLENOW FRAGMENT OF DNA POLYMERASE I, MAGNESIUM ION, ... | | Authors: | Brautigam, C.A, Sun, S, Piccirilli, J.A, Steitz, T.A. | | Deposit date: | 1998-07-01 | | Release date: | 1998-11-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of normal single-stranded DNA and deoxyribo-3'-S-phosphorothiolates bound to the 3'-5' exonucleolytic active site of DNA polymerase I from Escherichia coli.
Biochemistry, 38, 1999
|
|
2KZZ
 
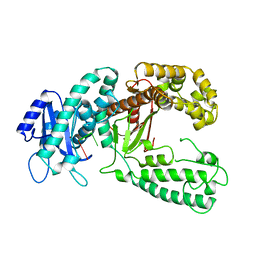 | | KLENOW FRAGMENT WITH NORMAL SUBSTRATE AND ZINC ONLY | | Descriptor: | DNA (5'-D(*GP*CP*TP*T*AP*CP*G)-3'), PROTEIN (DNA POLYMERASE I), ZINC ION | | Authors: | Brautigam, C.A, Sun, S, Piccirilli, J.A, Steitz, T.A. | | Deposit date: | 1998-07-07 | | Release date: | 1999-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of normal single-stranded DNA and deoxyribo-3'-S-phosphorothiolates bound to the 3'-5' exonucleolytic active site of DNA polymerase I from Escherichia coli.
Biochemistry, 38, 1999
|
|
2KZM
 
 | | KLENOW FRAGMENT WITH NORMAL SUBSTRATE AND ZINC AND MANGANESE | | Descriptor: | DNA (5'-D(*GP*CP*TP*TP*A*CP*GP*C)-3'), MANGANESE (II) ION, PROTEIN (DNA POLYMERASE I), ... | | Authors: | Brautigam, C.A, Sun, S, Piccirilli, J.A, Steitz, T.A. | | Deposit date: | 1998-07-03 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of normal single-stranded DNA and deoxyribo-3'-S-phosphorothiolates bound to the 3'-5' exonucleolytic active site of DNA polymerase I from Escherichia coli.
Biochemistry, 38, 1999
|
|
4KZD
 
 | | Crystal structure of an RNA aptamer in complex with fluorophore and Fab | | Descriptor: | 4-(3,5-difluoro-4-hydroxybenzyl)-1,2-dimethyl-1H-imidazol-5-ol, BL3-6 Fab antibody, heavy chain, ... | | Authors: | Huang, H, Suslov, N.B, Li, N, Koldobskaya, Y, Rice, P.A, Piccirilli, J.A. | | Deposit date: | 2013-05-29 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.186 Å) | | Cite: | A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore.
Nat.Chem.Biol., 10, 2014
|
|
4KZE
 
 | | Crystal structure of an RNA aptamer in complex with Fab | | Descriptor: | BL3-6 Fab antibody, heavy chain, light chain, ... | | Authors: | Huang, H, Suslov, N.B, Li, N, Koldobskaya, Y, Rice, P.A, Piccirilli, J.A. | | Deposit date: | 2013-05-29 | | Release date: | 2014-06-18 | | Last modified: | 2014-07-30 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore.
Nat.Chem.Biol., 10, 2014
|
|
7MLX
 
 | |
6R47
 
 | |
3HHN
 
 | | Crystal structure of class I ligase ribozyme self-ligation product, in complex with U1A RBD | | Descriptor: | Class I ligase ribozyme, self-ligation product, MAGNESIUM ION, ... | | Authors: | Shechner, D.M, Grant, R.A, Bagby, S.C, Bartel, D.P. | | Deposit date: | 2009-05-15 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.987 Å) | | Cite: | Crystal structure of the catalytic core of an RNA-polymerase ribozyme.
Science, 326, 2009
|
|
6MIH
 
 | | Crystal structure of host-guest complex with PC hachimoji DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*(1WA)P*CP*(DB)P*T)-3'), DNA (5'-D(P*AP*(DS)P*GP*(1W5)P*TP*AP*AP*G)-3'), N-terminal fragment of MMLV reverse transcriptase | | Authors: | Georgiadis, M.M. | | Deposit date: | 2018-09-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Hachimoji DNA and RNA: A genetic system with eight building blocks.
Science, 363, 2019
|
|
6MIG
 
 | | Crystal structure of host-guest complex with PB hachimoji DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*TP*(1WA)P*(1WA)P*(DS))-3'), DNA (5'-D(P*(DB)P*(1W5)P*(1W5)P*AP*TP*AP*AP*G)-3'), Gag-Pol polyprotein | | Authors: | Georgiadis, M.M. | | Deposit date: | 2018-09-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hachimoji DNA and RNA: A genetic system with eight building blocks.
Science, 363, 2019
|
|
6MIK
 
 | | Crystal structure of host-guest complex with PP hachimoji DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*TP*(1WA)P*(1WA)P*(DS))-3'), DNA (5'-D(P*(DB)P*(1W5)P*(1W5)P*AP*TP*AP*AP*G)-3'), N-terminal fragment of MMLV reverse transcriptase | | Authors: | Georgiadis, M.M. | | Deposit date: | 2018-09-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hachimoji DNA and RNA: A genetic system with eight building blocks.
Science, 363, 2019
|
|
3QHT
 
 | | Crystal Structure of the Monobody ySMB-1 bound to yeast SUMO | | Descriptor: | GLYCEROL, Monobody ySMB-1, Ubiquitin-like protein SMT3 | | Authors: | Koide, S, Gilbreth, R.N. | | Deposit date: | 2011-01-26 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Isoform-specific monobody inhibitors of small ubiquitin-related modifiers engineered using structure-guided library design.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7R6L
 
 | | 5 prime exon-free pre-2S intermediate of the Tetrahymena group I intron, symmetry-expanded monomer from a synthetic dimeric construct | | Descriptor: | Group I intron, 3 prime fragment plus 3 prime exon, 5 prime fragment, ... | | Authors: | Thelot, F, Liu, D, Liao, M, Yin, P. | | Deposit date: | 2021-06-22 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Sub-3- angstrom cryo-EM structure of RNA enabled by engineered homomeric self-assembly.
Nat.Methods, 19, 2022
|
|
7R6N
 
 | | Exon-free state of the Tetrahymena group I intron, symmetry-expanded monomer from a synthetic trimeric construct | | Descriptor: | Group I intron, MAGNESIUM ION | | Authors: | Thelot, F, Liu, D, Liao, M, Yin, P. | | Deposit date: | 2021-06-22 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Sub-3- angstrom cryo-EM structure of RNA enabled by engineered homomeric self-assembly.
Nat.Methods, 19, 2022
|
|
7R6M
 
 | | Post-2S intermediate of the Tetrahymena group I intron, symmetry-expanded monomer from a synthetic dimeric construct | | Descriptor: | Group I intron, Ligated exon mimic of the Group I intron, MAGNESIUM ION | | Authors: | Thelot, F, Liu, D, Liao, M, Yin, P. | | Deposit date: | 2021-06-22 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Sub-3- angstrom cryo-EM structure of RNA enabled by engineered homomeric self-assembly.
Nat.Methods, 19, 2022
|
|
4ONO
 
 | | CD1c in complex with PM (phosphomycoketide) | | Descriptor: | (4R,8S,16S,20R)-4,8,12,16,20-pentamethylheptacosyl dihydrogen phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin/T-cell surface glycoprotein CD1c/T-cell surface glycoprotein CD1b chimeric protein, ... | | Authors: | Roy, S, Adams, E.J. | | Deposit date: | 2014-01-28 | | Release date: | 2014-10-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Molecular basis of mycobacterial lipid antigen presentation by CD1c and its recognition by alpha beta T cells.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4ONH
 
 | | Crystal Structure of DN6 TCR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Roy, S, Adams, E.J. | | Deposit date: | 2014-01-28 | | Release date: | 2014-10-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.008 Å) | | Cite: | Molecular basis of mycobacterial lipid antigen presentation by CD1c and its recognition by alpha beta T cells.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
