6QGI
 
 | | Crystal structure of VP5 from Haloarchaeal pleomorphic virus 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, VP5 | | Authors: | El Omari, K, Walter, T.S, Harlos, K, Grimes, J.M, Stuart, D.I, Roine, E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The structure of a prokaryotic viral envelope protein expands the landscape of membrane fusion proteins.
Nat Commun, 10, 2019
|
|
6QGL
 
 | | Crystal structure of VP5 from Haloarchaeal pleomorphic virus 6 | | Descriptor: | BROMIDE ION, VP5 | | Authors: | El Omari, K, Walter, T.S, Harlos, K, Grimes, J.M, Stuart, D.I, Roine, E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | The structure of a prokaryotic viral envelope protein expands the landscape of membrane fusion proteins.
Nat Commun, 10, 2019
|
|
6JJH
 
 | | Crystal structure of a two-quartet RNA parallel G-quadruplex complexed with the porphyrin TMPyP4 | | Descriptor: | (1Z,4Z,9Z,15Z)-5,10,15,20-tetrakis(1-methylpyridin-1-ium-4-yl)-21,23-dihydroporphyrin, POTASSIUM ION, RNA (5'-R(*GP*GP*CP*UP*CP*GP*GP*CP*GP*GP*CP*GP*GP*A)-3') | | Authors: | Zhang, Y.S, EI Omari, K, Duman, R, Wagner, A, Parkinson, G.N, Wei, D.G. | | Deposit date: | 2019-02-25 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths.
Nucleic Acids Res., 48, 2020
|
|
6JJF
 
 | | Crystal structure of a two-quartet DNA mixed-parallel/antiparallel G-quadruplex | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*GP*GP*CP*TP*CP*GP*GP*CP*GP*GP*CP*GP*GP*A)-3'), POTASSIUM ION, ... | | Authors: | Zhang, Y.S, EI Omari, K, Duman, R, Wagner, A, Parkinson, G.N, Wei, D.G. | | Deposit date: | 2019-02-25 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths.
Nucleic Acids Res., 48, 2020
|
|
7ACX
 
 | | H/L (SLPH/SLPL) complex from C. difficile (R7404 strain) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S-layer protein, SULFATE ION | | Authors: | Lanzoni-Mangutchi, P, Barwinska-Sendra, A, Basle, A, El Omari, K, Wagner, A, Salgado, P.S. | | Deposit date: | 2020-09-11 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and assembly of the S-layer in C. difficile.
Nat Commun, 13, 2022
|
|
7QE2
 
 | | Crystal structure of D-glucuronic acid bound to SN243 | | Descriptor: | ACETATE ION, SN243, SULFATE ION, ... | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
7QG4
 
 | | Apo crystal structure of a mutant of SN243 (D415N) | | Descriptor: | SN243, SULFATE ION, ZINC ION | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-07 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
7QEF
 
 | | Crystal structure of para-nitrophenyl-Beta-D-glucuronide bound to a mutant of SN243 (D415A) | | Descriptor: | 4-nitrophenyl beta-D-glucopyranosiduronic acid, ACETATE ION, SN243, ... | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-02 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
7QEA
 
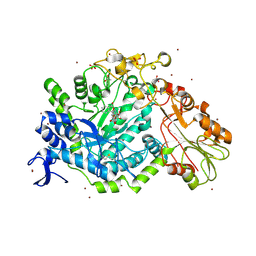 | | Crystal structure of fluorescein-di-Beta-D-glucuronide bound to a mutant of SN243 (D415A) | | Descriptor: | (2~{S},3~{S},4~{S},5~{R},6~{S})-3,4,5-tris(oxidanyl)-6-[(1~{R})-6'-oxidanyl-3-oxidanylidene-spiro[2-benzofuran-1,9'-xanthene]-3'-yl]oxy-oxane-2-carboxylic acid, ACETATE ION, SN243, ... | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
7QE1
 
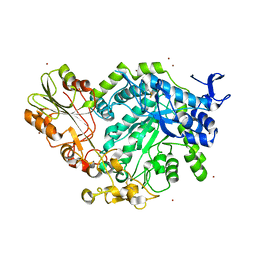 | | Crystal structure of apo SN243 | | Descriptor: | SN243, ZINC ION | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-12 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
7QEE
 
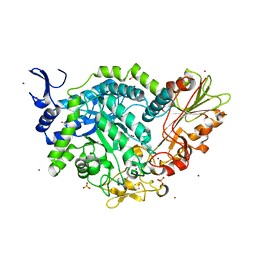 | | SN243 mutant D415N bound to para-nitrophenyl-Beta-D-glucuronide | | Descriptor: | 4-nitrophenyl beta-D-glucopyranosiduronic acid, SN243, SULFATE ION, ... | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-02 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.374 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
6J7V
 
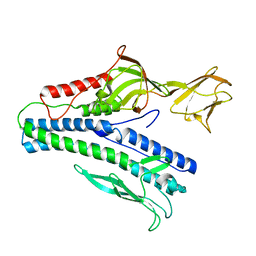 | |
8CI3
 
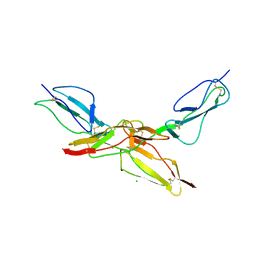 | | Structure of bovine CD46 ectodomain (SCR 1-2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Aitkenhead, H, David I Stuart, D.I, El Omari, K. | | Deposit date: | 2023-02-08 | | Release date: | 2023-07-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure of Bovine CD46 Ectodomain.
Viruses, 15, 2023
|
|
8CJV
 
 | |
4BX4
 
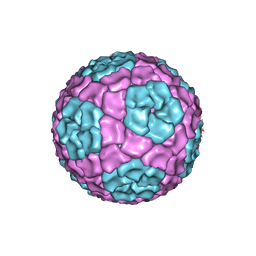 | | Fitting of the bacteriophage Phi8 P1 capsid protein into cryo-EM density | | Descriptor: | P1 | | Authors: | El Omari, K, Sutton, G, Ravantti, J.J, Zhang, H, Walter, T.S, Grimes, J.M, Bamford, D.H, Stuart, D.I, Mancini, E.J. | | Deposit date: | 2013-07-08 | | Release date: | 2013-08-07 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Plate Tectonics of Virus Shell Assembly and Reorganization in Phage Phi8, a Distant Relative of Mammalian Reoviruses
Structure, 21, 2013
|
|
4BLS
 
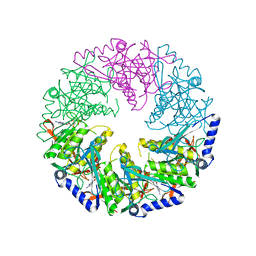 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 Q278A MUTANT IN COMPLEX WITH AMPcPP | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, NTPASE P4 | | Authors: | El Omari, K, Meier, C, Kainov, D, Sutton, G, Grimes, J.M, Poranen, M.M, Bamford, D.H, Tuma, R, Stuart, D.I, Mancini, E.J. | | Deposit date: | 2013-05-04 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Tracking in Atomic Detail the Functional Specializations in Viral Reca Helicases that Occur During Evolution.
Nucleic Acids Res., 41, 2013
|
|
4BLT
 
 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 S292A MUTANT IN COMPLEX WITH AMPcPP | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, NTPASE P4 | | Authors: | El Omari, K, Meier, C, Kainov, D, Sutton, G, Grimes, J.M, Poranen, M.M, Bamford, D.H, Tuma, R, Stuart, D.I, Mancini, E.J. | | Deposit date: | 2013-05-04 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Tracking in Atomic Detail the Functional Specializations in Viral Reca Helicases that Occur During Evolution.
Nucleic Acids Res., 41, 2013
|
|
4BLR
 
 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 IN COMPLEX WITH UTP | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, NTPASE P4, URIDINE 5'-TRIPHOSPHATE | | Authors: | El Omari, K, Meier, C, Kainov, D, Sutton, G, Grimes, J.M, Poranen, M.M, Bamford, D.H, Tuma, R, Stuart, D.I, Mancini, E.J. | | Deposit date: | 2013-05-04 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tracking in Atomic Detail the Functional Specializations in Viral Reca Helicases that Occur During Evolution.
Nucleic Acids Res., 41, 2013
|
|
4BWY
 
 | | P4 PROTEIN FROM BACTERIOPHAGE PHI8 (R32) | | Descriptor: | P4 | | Authors: | El Omari, K, Meier, C, Kainov, D, Sutton, G, Grimes, J.M, Poranen, M.M, Bamford, D.H, Tuma, R, Stuart, D.I, Mancini, E.J. | | Deposit date: | 2013-07-05 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Tracking in Atomic Detail the Functional Specializations in Viral Reca Helicases that Occur During Evolution.
Nucleic Acids Res., 41, 2013
|
|
4BTP
 
 | | Structure of the capsid protein P1 of the bacteriophage phi8 | | Descriptor: | p1 | | Authors: | El Omari, K, Sutton, G, Ravantti, J.J, Zhang, H, Walter, T.S, Grimes, J.M, Bamford, D.H, Stuart, D.I, Mancini, E.J. | | Deposit date: | 2013-06-18 | | Release date: | 2013-08-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Plate Tectonics of Virus Shell Assembly and Reorganization in Phage Phi8, a Distant Relative of Mammalian Reoviruses
Structure, 21, 2013
|
|
8PX0
 
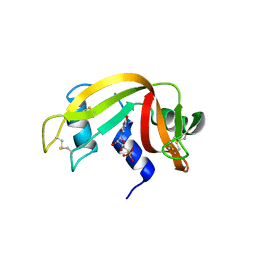 | | Structure of ribonuclease A, solved at wavelength 2.75 A | | Descriptor: | L-URIDINE-5'-MONOPHOSPHATE, Ribonuclease pancreatic | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Romano, M, Beis, K, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PX7
 
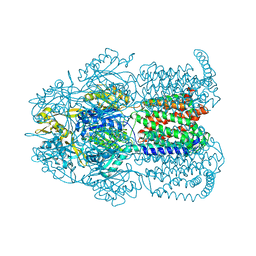 | | Structure of Bacterial Multidrug Efflux transporter AcrB, solved at wavelength 3.02 A | | Descriptor: | Multidrug efflux pump subunit AcrB | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Qu, F, Beis, K, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PX9
 
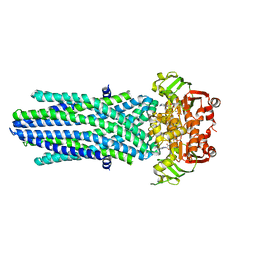 | | Structure of the antibacterial peptide ABC transporter McjD, solved at wavelength 2.75 A | | Descriptor: | MAGNESIUM ION, Microcin-J25 export ATP-binding/permease protein McjD, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Bountra, K, Beis, K, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PXH
 
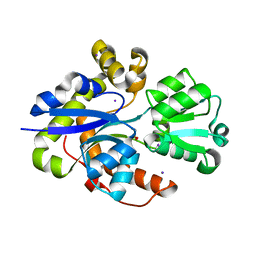 | | Structure of TauA from E. coli, solved at wavelength 2.375 A | | Descriptor: | 2-AMINOETHANESULFONIC ACID, IODIDE ION, Taurine ABC transporter substrate-binding protein | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Qu, F, Beis, K, Wagner, A. | | Deposit date: | 2023-07-23 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PYZ
 
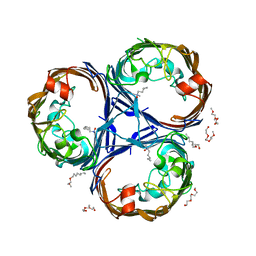 | | Structure of Ompk36GD from Klebsiella pneumonia, solved at wavelength 4.13 A | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, LAURYL DIMETHYLAMINE-N-OXIDE, OmpK36 | | Authors: | Duman, R, El Omari, K, Mykhaylyk, V, Orr, C, Kwong, H, Beis, K, Wagner, A. | | Deposit date: | 2023-07-26 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
