2IHS
 
 | |
6IOZ
 
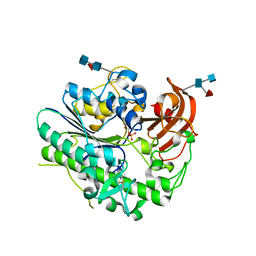 | | Structural insights of idursulfase beta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kim, H, Kim, D, Hong, J, Lee, K, Seo, J, Oh, B.H. | | Deposit date: | 2018-11-01 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights of idursulfase beta
To Be Published
|
|
4DWZ
 
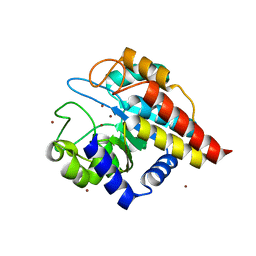 | | Crystal Structure of Ton_0340 | | Descriptor: | Hypothetical protein TON_0340, ZINC ION | | Authors: | Lee, S.G, Lee, K.H, Cha, S.S, Oh, B.H. | | Deposit date: | 2012-02-27 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Experimental phasing using zinc anomalous scattering
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4EOG
 
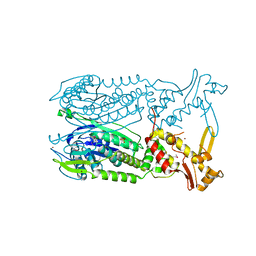 | | Crystal structure of Csx1 of Pyrococcus furiosus | | Descriptor: | Putative uncharacterized protein, SULFATE ION, ZINC ION | | Authors: | Kim, Y.K, Oh, B.H. | | Deposit date: | 2012-04-14 | | Release date: | 2013-01-02 | | Last modified: | 2013-07-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and nucleic acid-binding activity of the CRISPR-associated protein Csx1 of Pyrococcus furiosus.
Proteins, 81, 2013
|
|
4L6U
 
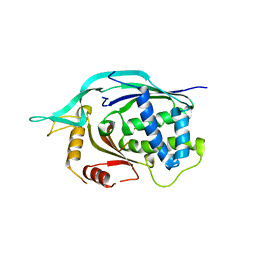 | | Crystal structure of AF1868: Cmr1 subunit of the Cmr RNA silencing complex | | Descriptor: | Putative uncharacterized protein | | Authors: | Sun, J, Jeon, J.H, Shin, M, Shin, H.C, Oh, B.H, Kim, J.S. | | Deposit date: | 2013-06-12 | | Release date: | 2014-02-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and CRISPR RNA-binding site of the Cmr1 subunit of the Cmr interference complex
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2WWX
 
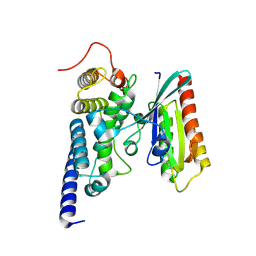 | | Crystal structure of the SidM/DrrA(GEF/GDF domain)-Rab1(GTPase domain) complex | | Descriptor: | DRRA, RAS-RELATED PROTEIN RAB-1 | | Authors: | Suh, H.Y, Lee, D.W, Woo, J.S, Oh, B.H. | | Deposit date: | 2009-10-30 | | Release date: | 2009-12-08 | | Last modified: | 2015-04-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights Into the Dual Nucleotide Exchange and Gdi Displacement Activity of Sidm/Drra
Embo J., 29, 2010
|
|
2WP7
 
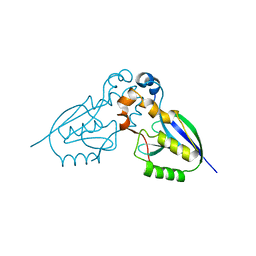 | | Crystal structure of deSUMOylase(DUF862) | | Descriptor: | PPPDE PEPTIDASE DOMAIN-CONTAINING PROTEIN 2 | | Authors: | Kim, J.H, Woo, J.S, Oh, B.H. | | Deposit date: | 2009-08-03 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Desi-1, a Novel Desumoylase Belonging to a Putative Isopeptidase Superfamily.
Proteins, 80, 2012
|
|
2XA0
 
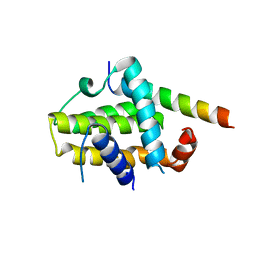 | | Crystal structure of BCL-2 in complex with a BAX BH3 peptide | | Descriptor: | APOPTOSIS REGULATOR BAX, APOPTOSIS REGULATOR BCL-2 | | Authors: | Ku, B, Oh, B.H. | | Deposit date: | 2010-03-25 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Evidence that Inhibition of Bax Activation by Bcl- 2 Involves its Tight and Preferential Interaction with the Bh3 Domain of Bax.
Cell Res., 21, 2011
|
|
1K41
 
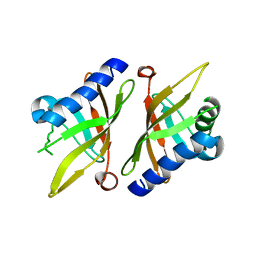 | | Crystal structure of KSI Y57S mutant | | Descriptor: | Ketosteroid Isomerase | | Authors: | Cha, S.S, Oh, B.H, Nam, G.H, Jang, D.S, Lee, T.H, Choi, K.Y. | | Deposit date: | 2001-10-05 | | Release date: | 2002-10-16 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Maintenance of alpha-helical structures by phenyl rings in the active-site tyrosine triad contributes to catalysis and stability of ketosteroid isomerase from Pseudomonas putida biotype B
Biochemistry, 40, 2001
|
|
5X1E
 
 | |
5X1U
 
 | |
5X1H
 
 | | Structure of Legionella pneumophila DotN | | Descriptor: | IcmJ (DotN), ZINC ION | | Authors: | Kwak, M.J, Oh, B.H. | | Deposit date: | 2017-01-26 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Architecture of the type IV coupling protein complex of Legionella pneumophila
Nat Microbiol, 2, 2017
|
|
3EUJ
 
 | | Crystal structure of MukE-MukF(residues 292-443)-MukB(head domain)-ATPgammaS complex, symmetric dimer | | Descriptor: | Chromosome partition protein mukB, Linker, Chromosome partition protein mukF, ... | | Authors: | Woo, J.S, Lim, J.H, Shin, H.C, Oh, B.H. | | Deposit date: | 2008-10-10 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions.
Cell(Cambridge,Mass.), 136, 2009
|
|
3EUK
 
 | | Crystal structure of MukE-MukF(residues 292-443)-MukB(head domain)-ATPgammaS complex, asymmetric dimer | | Descriptor: | Chromosome partition protein mukB, Linker, Chromosome partition protein mukE, ... | | Authors: | Woo, J.S, Lim, J.H, Shin, H.C, Oh, B.H. | | Deposit date: | 2008-10-10 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions.
Cell(Cambridge,Mass.), 136, 2009
|
|
3EUH
 
 | | Crystal Structure of the MukE-MukF Complex | | Descriptor: | Chromosome partition protein mukF, GLYCINE, MukE | | Authors: | Suh, M.K, Ku, B, Ha, N.C, Woo, J.S, Oh, B.H. | | Deposit date: | 2008-10-10 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions.
Cell(Cambridge,Mass.), 136, 2009
|
|
4FC5
 
 | | Crystal Structure of Ton_0340 | | Descriptor: | Putative uncharacterized protein, ZINC ION | | Authors: | Lee, S.G, Lee, K.H, An, Y.J, Cha, S.S, Oh, B.H. | | Deposit date: | 2012-05-24 | | Release date: | 2012-09-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Experimental phasing using zinc anomalous scattering
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4I99
 
 | |
4I98
 
 | |
4JGH
 
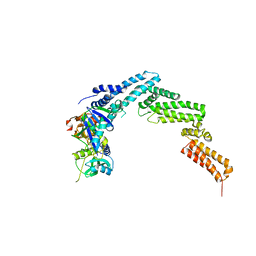 | | Structure of the SOCS2-Elongin BC complex bound to an N-terminal fragment of Cullin5 | | Descriptor: | Cullin-5, Suppressor of cytokine signaling 2, Transcription elongation factor B polypeptide 1, ... | | Authors: | Kim, Y.K, Kwak, M.J, Ku, B, Suh, H.Y, Joo, K, Lee, J, Jung, J.U, Oh, B.H. | | Deposit date: | 2013-03-01 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of intersubunit recognition in elongin BC-cullin 5-SOCS box ubiquitin-protein ligase complexes.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5X42
 
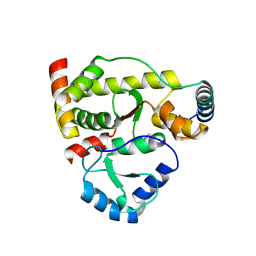 | | Structure of DotL(590-659)-DotN derived from Legionella pneumophila | | Descriptor: | DotN, IcmJ (DotN), IcmO (DotL), ... | | Authors: | Kwak, M.J, Kim, J.D, Oh, B.H. | | Deposit date: | 2017-02-09 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Architecture of the type IV coupling protein complex of Legionella pneumophila
Nat Microbiol, 2, 2017
|
|
5X90
 
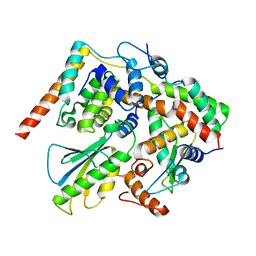 | | Structure of DotL(656-783)-IcmS-IcmW-LvgA derived from Legionella pneumophila | | Descriptor: | Hypothetical virulence protein, IcmO (DotL), IcmS, ... | | Authors: | Kim, H, Kwak, M.J, Kim, J.D, Kim, Y.G, Oh, B.H. | | Deposit date: | 2017-03-04 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Architecture of the type IV coupling protein complex of Legionella pneumophila
Nat Microbiol, 2, 2017
|
|
2LAO
 
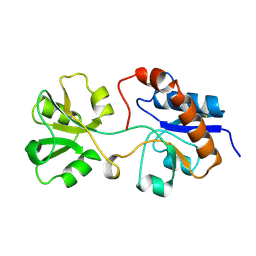 | | THREE-DIMENSIONAL STRUCTURES OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN WITH AND WITHOUT A LIGAND | | Descriptor: | LYSINE, ARGININE, ORNITHINE-BINDING PROTEIN | | Authors: | Kim, S.-H, Oh, B.-H, Kang, C.-H. | | Deposit date: | 1993-02-25 | | Release date: | 1994-06-22 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-dimensional structures of the periplasmic lysine/arginine/ornithine-binding protein with and without a ligand.
J.Biol.Chem., 268, 1993
|
|
1AYP
 
 | |
1LAH
 
 | | STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN | | Descriptor: | L-ornithine, LYSINE, ARGININE, ... | | Authors: | Kim, S.-H, Oh, B.-H. | | Deposit date: | 1993-10-06 | | Release date: | 1995-07-10 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein.
J.Biol.Chem., 269, 1994
|
|
1LAF
 
 | | STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN | | Descriptor: | ARGININE, LYSINE, ORNITHINE-BINDING PROTEIN | | Authors: | Kim, S.-H, Oh, B.-H. | | Deposit date: | 1993-10-06 | | Release date: | 1995-07-10 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein.
J.Biol.Chem., 269, 1994
|
|
