1RGX
 
 | | Crystal Structure of resisitin | | Descriptor: | Resistin | | Authors: | Patel, S.D, Rajala, M.W, Scherer, P.E, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-11-13 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.787 Å) | | Cite: | Disulfide-dependent multimeric assembly of resistin family hormones
Science, 304, 2004
|
|
1S7J
 
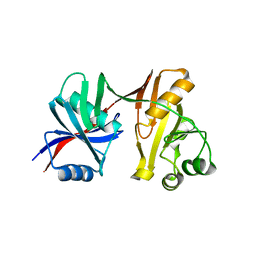 | |
1RZN
 
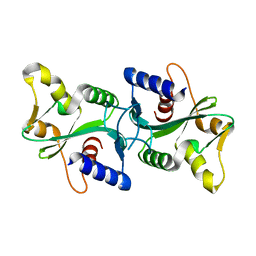 | |
1YRZ
 
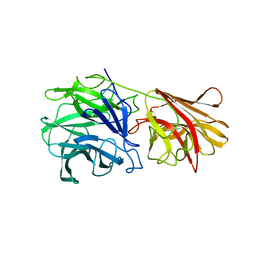 | | Crystal structure of xylan beta-1,4-xylosidase from Bacillus Halodurans C-125 | | Descriptor: | xylan beta-1,4-xylosidase | | Authors: | Fedorov, A.A, Fedorov, E.V, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-02-05 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of xylan beta-1,4-xylosidase from Bacillus Halodurans C-125
To be Published
|
|
1YMY
 
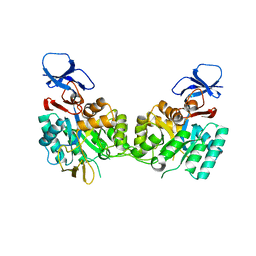 | | Crystal Structure of the N-Acetylglucosamine-6-phosphate deacetylase from Escherichia coli K12 | | Descriptor: | N-acetylglucosamine-6-phosphate deacetylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Xiang, D.F, Raushel, F.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-01-21 | | Release date: | 2005-02-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the N-acetylglucosamine-6-phosphate deacetylase from Escherichia Coli
To be Published
|
|
1YWK
 
 | |
1Z4E
 
 | |
3S6J
 
 | | The crystal structure of a hydrolase from Pseudomonas syringae | | Descriptor: | CALCIUM ION, Hydrolase, haloacid dehalogenase-like family | | Authors: | Zhang, Z, Syed Ibrahim, B, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-05-25 | | Release date: | 2011-07-13 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | The crystal structure of a hydrolase from Pseudomonas syringae
To be Published
|
|
3SLR
 
 | | Crystal structure of N-terminal part of the protein BF1531 from Bacteroides fragilis containing phosphatase domain complexed with Mg. | | Descriptor: | MAGNESIUM ION, uncharacterized protein BF1531 | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-06-24 | | Release date: | 2011-07-20 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Crystal structure of N-terminal part of the protein BF1531 from Bacteroides fragilis
containing phosphatase domain complexed with Mg.
TO BE PUBLISHED
|
|
2A1V
 
 | |
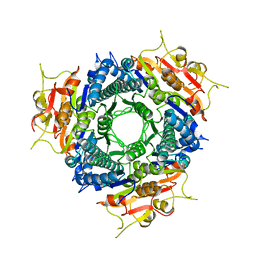
2A1F
 
 | |
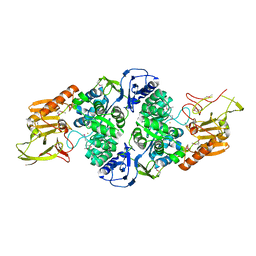
3T81
 
 | |
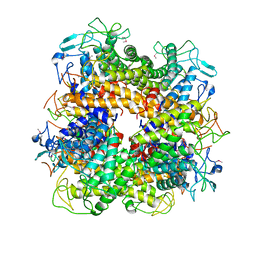
2AFA
 
 | |
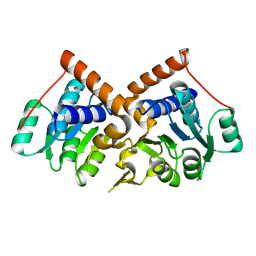
1PQW
 
 | |
2P27
 
 | | Crystal Structure of Human Pyridoxal Phosphate Phosphatase with Mg2+ at 1.9 A resolution | | Descriptor: | MAGNESIUM ION, Pyridoxal phosphate phosphatase | | Authors: | Ramagopal, U.A, Freeman, J, Izuka, M, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-07 | | Release date: | 2007-03-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
1XM5
 
 | | Crystal structure of metal-dependent hydrolase ybeY from E. coli, Pfam UPF0054 | | Descriptor: | Hypothetical UPF0054 protein ybeY, NICKEL (II) ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Shi, W, Ramagopal, U.A, Thirumuruhan, R, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-10-01 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The ybeY protein from Escherichia coli is a metalloprotein.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
2F7F
 
 | | Crystal structure of Enterococcus faecalis putative nicotinate phosphoribosyltransferase, NEW YORK STRUCTURAL GENOMICS CONSORTIUM | | Descriptor: | DIPHOSPHATE, GLYCEROL, NICOTINIC ACID, ... | | Authors: | Patskovsky, Y, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-11-30 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Enterococcus Faecalis Nicotinate Phosphoribosyltransferase
To be Published
|
|
3SMD
 
 | |
1YIF
 
 | | CRYSTAL STRUCTURE OF beta-1,4-xylosidase FROM BACILLUS SUBTILIS, NEW YORK STRUCTURAL GENOMICS CONSORTIUM | | Descriptor: | BETA-1,4-XYLOSIDASE | | Authors: | Patskovsky, Y, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-01-11 | | Release date: | 2005-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF beta-1,4-xylosidase FROM BACILLUS SUBTILIS
To be Published
|
|
3R03
 
 | |
2HSI
 
 | |
1YGA
 
 | |
1Y9I
 
 | | Crystal structure of low temperature requirement C protein from Listeria monocytogenes | | Descriptor: | CALCIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-15 | | Release date: | 2004-12-28 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of phosphatidylglycerophosphatase (PGPase), a putative membrane-bound lipid phosphatase, reveals a novel binuclear metal binding site and two "proton wires".
Proteins, 64, 2006
|
|
2OYC
 
 | | Crystal structure of human pyridoxal phosphate phosphatase | | Descriptor: | Pyridoxal phosphate phosphatase, SODIUM ION, TUNGSTATE(VI)ION | | Authors: | Ramagopal, U.A, Freeman, J, Izuka, M, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-21 | | Release date: | 2007-03-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
2P69
 
 | | Crystal Structure of Human Pyridoxal Phosphate Phosphatase with PLP | | Descriptor: | CALCIUM ION, PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate phosphatase | | Authors: | Ramagopal, U.A, Freeman, J, Izuka, M, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
