1JZT
 
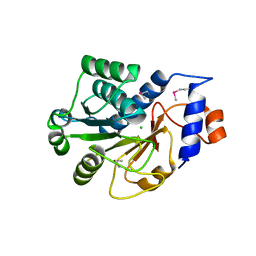 | |
1K4Z
 
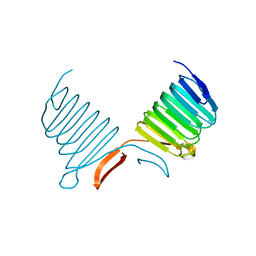 | | C-terminal Domain of Cyclase Associated Protein | | Descriptor: | Adenylyl Cyclase-Associated Protein | | Authors: | Rozwarski, D.A, Fedorov, A.A, Dodatko, T, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-10-09 | | Release date: | 2002-03-13 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the actin binding domain of the cyclase-associated protein
Biochemistry, 43, 2004
|
|
1K8F
 
 | | CRYSTAL STRUCTURE OF THE HUMAN C-TERMINAL CAP1-ADENYLYL CYCLASE ASSOCIATED PROTEIN | | Descriptor: | ADENYLYL CYCLASE-ASSOCIATED PROTEIN | | Authors: | Patskovsky, Y.V, Chance, M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-10-24 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the actin binding domain of the cyclase-associated protein.
Biochemistry, 43, 2004
|
|
1KCX
 
 | | X-ray structure of NYSGRC target T-45 | | Descriptor: | DIHYDROPYRIMIDINASE RELATED PROTEIN-1 | | Authors: | Deo, R.C, Schmidt, E.F, Strittmatter, S.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-11-11 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural bases for CRMP function in plexin-dependent semaphorin3A signaling
Embo J., 23, 2004
|
|
1KU9
 
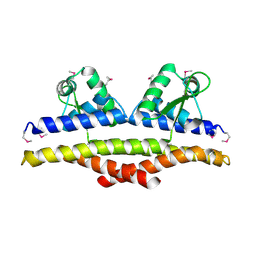 | | X-ray Structure of a Methanococcus jannaschii DNA-Binding Protein: Implications for Antibiotic Resistance in Staphylococcus aureus | | Descriptor: | hypothetical protein MJ223 | | Authors: | Ray, S.S, Bonanno, J.B, Chen, H, de Lencastre, H, Wu, S, Tomasz, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-01-21 | | Release date: | 2002-12-25 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of an M. jannaschii DNA-binding protein: implications for antibiotic resistance in S.
aureus
Proteins, 50, 2002
|
|
1KMK
 
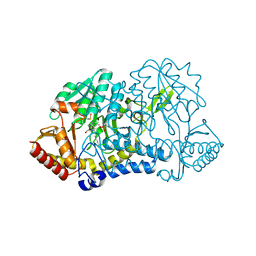 | |
1KMJ
 
 | |
1M0T
 
 | |
4F2D
 
 | | Crystal Structure of Escherichia coli L-arabinose Isomerase (ECAI) complexed with Ribitol | | Descriptor: | ACETIC ACID, D-ribitol, L-arabinose isomerase, ... | | Authors: | Manjasetty, B.A, Burley, S.K, Almo, S.C, Chance, M.R, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2012-05-07 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Escherichia coli L-arabinose Isomerase (ECAI) complexed with Ribitol
TO BE PUBLISHED
|
|
1HQZ
 
 | | Cofilin homology domain of a yeast actin-binding protein ABP1P | | Descriptor: | ACTIN-BINDING PROTEIN | | Authors: | Strokopytov, B.V, Fedorov, A.A, Mahoney, N, Drubin, D.G, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2000-12-20 | | Release date: | 2001-12-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phased translation function revisited: structure solution of the cofilin-homology domain from yeast actin-binding protein 1 using six-dimensional searches.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2HCM
 
 | | Crystal structure of mouse putative dual specificity phosphatase complexed with zinc tungstate, New York Structural Genomics Consortium | | Descriptor: | Dual specificity protein phosphatase, GLYCEROL, SODIUM ION, ... | | Authors: | Patskovsky, Y, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-06-17 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
1JFI
 
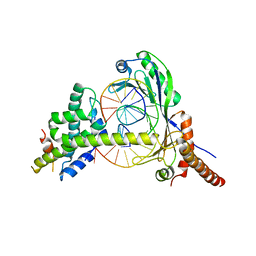 | | Crystal Structure of the NC2-TBP-DNA Ternary Complex | | Descriptor: | 5'-D(*G*GP*AP*GP*CP*CP*CP*TP*TP*TP*TP*AP*TP*AP*GP*CP*CP*AP*A)-3', 5'-D(*TP*TP*GP*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*GP*CP*TP*CP*C)-3', TATA-BOX-BINDING PROTEIN (TBP), ... | | Authors: | Kamada, K, Shu, F, Chen, H, Malik, S, Stelzer, G, Roeder, R.G, Meisterernst, M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-06-20 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structure of negative cofactor 2 recognizing the TBP-DNA transcription complex.
Cell(Cambridge,Mass.), 106, 2001
|
|
1JSS
 
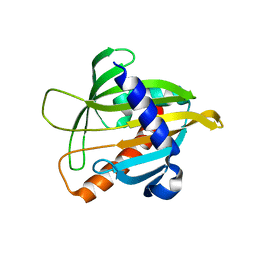 | | Crystal structure of the Mus musculus cholesterol-regulated START protein 4 (StarD4). | | Descriptor: | cholesterol-regulated START protein 4 | | Authors: | Romanowski, M.J, Soccio, R.E, Breslow, J.L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-08-17 | | Release date: | 2002-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Mus musculus cholesterol-regulated START protein 4 (StarD4) containing a StAR-related lipid transfer domain.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1K47
 
 | |
1L9G
 
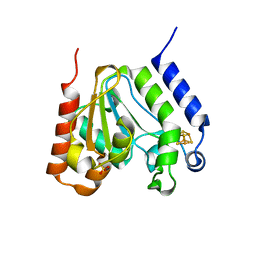 | | CRYSTAL STRUCTURE OF URACIL-DNA GLYCOSYLASE FROM T. MARITIMA | | Descriptor: | Conserved hypothetical protein, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Rajashankar, K.R, Dodatko, T, Thirumuruhan, R.A, Sandigursky, M, Bresnik, A, Chance, M.R, Franklin, W.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-03-22 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of uracil-DNA glycosylase from T. Maritima
To be Published
|
|
1LX7
 
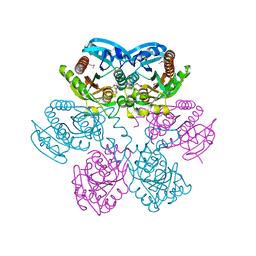 | | Structure of E. coli uridine phosphorylase at 2.0A | | Descriptor: | uridine phosphorylase | | Authors: | Burling, T, Buglino, J.A, Kniewel, R, Chadna, T, Beckwith, A, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-06-04 | | Release date: | 2002-06-12 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Escherichia coli uridine phosphorylase at 2.0 A.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1Z2I
 
 | |
1JD1
 
 | |
1JYH
 
 | |
1JSX
 
 | |
1KAG
 
 | |
2ISN
 
 | | Crystal structure of a phosphatase from a pathogenic strain Toxoplasma gondii | | Descriptor: | NYSGXRC-8828z, phosphatase, PRASEODYMIUM ION, ... | | Authors: | Agarwal, R, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-18 | | Release date: | 2006-10-31 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2AA4
 
 | |
2HHL
 
 | | Crystal structure of the human small CTD phosphatase 3 isoform 1 | | Descriptor: | 12-TUNGSTOPHOSPHATE, CTD small phosphatase-like protein | | Authors: | Malashkevich, V.N, Toro, R, Ramagopal, U, Sauder, J.M, Schwinn, K.D, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-06-28 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2FH7
 
 | | Crystal structure of the phosphatase domains of human PTP SIGMA | | Descriptor: | Receptor-type tyrosine-protein phosphatase S | | Authors: | Alvarado, J, Udupi, R, Smith, D, Koss, J, Wasserman, S.R, Ozyurt, S, Atwell, S, Powell, A, Kearins, M.C, Rooney, I, Maletic, M, Bain, K.T, Freeman, J.C, Russell, M, Thompson, D.A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-12-23 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
