1X2H
 
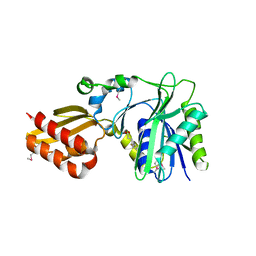 | | Crystal Structure of Lipate-Protein Ligase A from Escherichia coli complexed with lipoic acid | | Descriptor: | LIPOIC ACID, Lipoate-protein ligase A | | Authors: | Fujiwara, K, Toma, S, Okamura-Ikeda, K, Motokawa, Y, Nakagawa, A, Taniguchi, H. | | Deposit date: | 2005-04-23 | | Release date: | 2005-08-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structure of lipoate-protein ligase A from Escherichia coli: Determination of the lipoic acid-binding site
J.Biol.Chem., 280, 2005
|
|
1X2G
 
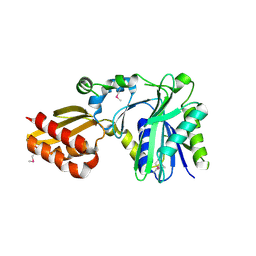 | | Crystal Structure of Lipate-Protein Ligase A from Escherichia coli | | Descriptor: | Lipoate-protein ligase A | | Authors: | Fujiwara, K, Toma, S, Okamura-Ikeda, K, Motokawa, Y, Nakagawa, A, Taniguchi, H. | | Deposit date: | 2005-04-23 | | Release date: | 2005-08-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of lipoate-protein ligase A from Escherichia coli: Determination of the lipoic acid-binding site
J.Biol.Chem., 280, 2005
|
|
2DE0
 
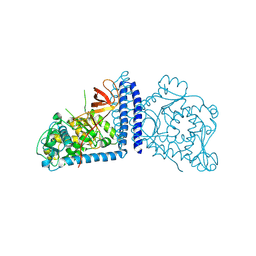 | | Crystal structure of human alpha 1,6-fucosyltransferase, FUT8 | | Descriptor: | Alpha-(1,6)-fucosyltransferase | | Authors: | Taniguchi, N, Ihara, H, Nakagawa, A. | | Deposit date: | 2006-02-07 | | Release date: | 2006-12-26 | | Last modified: | 2020-03-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of mammalian {alpha}1,6-fucosyltransferase, FUT8
Glycobiology, 17, 2007
|
|
1Y43
 
 | | crystal structure of aspergilloglutamic peptidase from Aspergillus niger | | Descriptor: | Aspergillopepsin II heavy chain, Aspergillopepsin II light chain, SULFATE ION | | Authors: | Sasaki, H, Nakagawa, A, Iwata, S, Muramatsu, T, Suganuma, M, Sawano, Y, Kojima, M, Kubota, K, Takahashi, K. | | Deposit date: | 2004-11-30 | | Release date: | 2005-12-13 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The three-dimensional structure of aspergilloglutamic peptidase from Aspergillus niger
Proc.Jpn.Acad.,Ser.B, 80, 2004
|
|
5WY1
 
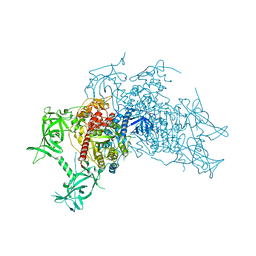 | | Crystal structure of mouse DNA methyltransferase 1 (T1505A mutant) | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, ZINC ION | | Authors: | Kanada, K, Takeshita, K, Suetake, I, Tajima, S, Nakagawa, A. | | Deposit date: | 2017-01-10 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Conserved threonine 1505 in the catalytic domain stabilizes mouse DNA methyltransferase 1
J. Biochem., 162, 2017
|
|
5YVQ
 
 | | Complex of Mu phage tail fiber and its chaperone | | Descriptor: | GLYCEROL, Tail fiber assembly protein U, Tail fiber protein S | | Authors: | Takeda, S, Sakai, K, Iwazaki, T, Yamashita, E, Nakagawa, A. | | Deposit date: | 2017-11-27 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Phage tail fibre assembly proteins employ a modular structure to drive the correct folding of diverse fibres.
Nat Microbiol, 4, 2019
|
|
5Z1N
 
 | | Crystal structure of C terminal region of G-protein interacting protein 1 (Gip1) from Dictyostelium discoideum | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, G-protein interacting protein 1, ... | | Authors: | Miyagawa, T, Koteishi, H, Kamimura, Y, Miyanaga, Y, Takeshita, K, Nakagawa, A, Ueda, M. | | Deposit date: | 2017-12-27 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Structural basis of Gip1 for cytosolic sequestration of G protein in wide-range chemotaxis
Nat Commun, 9, 2018
|
|
5Z39
 
 | | Crystal structure of C terminal region of G-protein interacting protein 1 (Gip1) from Dictyostelium discoideum form II | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, G-protein interacting protein 1, ... | | Authors: | Miyagawa, T, Koteishi, H, Kamimura, Y, Miyanaga, Y, Takeshita, K, Nakagawa, A, Ueda, M. | | Deposit date: | 2018-01-05 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural basis of Gip1 for cytosolic sequestration of G protein in wide-range chemotaxis
Nat Commun, 9, 2018
|
|
1UHI
 
 | | Crystal structure of i-aequorin | | Descriptor: | (2R)-8-BENZYL-2-HYDROPEROXY-6-(4-HYDROXYPHENYL)-2-(4-IODOBENZYL)-7,8-DIHYDROIMIDAZO[1,2-A]PYRAZIN-3(2H)-ONE, Aequorin 2 | | Authors: | Toma, S, Chong, K.T, Nakagawa, A, Teranishi, K, Inouye, S, Shimomura, O. | | Deposit date: | 2003-07-03 | | Release date: | 2005-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structures of semi-synthetic aequorins
Protein Sci., 14, 2005
|
|
1UHH
 
 | | Crystal structure of cp-aequorin | | Descriptor: | (8R)-8-(CYCLOPENTYLMETHYL)-2-HYDROPEROXY-2-(4-HYDROXYBENZYL)-6-(4-HYDROXYPHENYL)-7,8-DIHYDROIMIDAZO[1,2-A]PYRAZIN-3(2H) -ONE, Aequorin 2 | | Authors: | Toma, S, Chong, K.T, Nakagawa, A, Teranishi, K, Inouye, S, Shimomura, O. | | Deposit date: | 2003-07-03 | | Release date: | 2005-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structures of semi-synthetic aequorins
Protein Sci., 14, 2005
|
|
1UHJ
 
 | | Crystal structure of br-aequorin | | Descriptor: | (2S,8R)-8-BENZYL-2-(4-BROMOBENZYL)-2-HYDROPEROXY-6-(4-HYDROXYPHENYL)-7,8-DIHYDROIMIDAZO[1,2-A]PYRAZIN-3(2H)-ONE, Aequorin 2 | | Authors: | Toma, S, Chong, K.T, Nakagawa, A, Teranishi, K, Inouye, S, Shimomura, O. | | Deposit date: | 2003-07-03 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structures of semi-synthetic aequorins
Protein Sci., 14, 2005
|
|
1UHK
 
 | | Crystal structure of n-aequorin | | Descriptor: | (2S,8R)-8-BENZYL-2-HYDROPEROXY-6-(4-HYDROXYPHENYL)-2-(2-NAPHTHYLMETHYL)-7,8-DIHYDROIMIDAZO[1,2-A]PYRAZIN-3(2H)-ONE, Aequorin 2 | | Authors: | Toma, S, Chong, K.T, Nakagawa, A, Teranishi, K, Inouye, S, Shimomura, O. | | Deposit date: | 2003-07-03 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structures of semi-synthetic aequorins
Protein Sci., 14, 2005
|
|
3A7A
 
 | | Crystal structure of E. coli lipoate-protein ligase A in complex with octyl-amp and apoH-protein | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glycine cleavage system H protein, Lipoate-protein ligase A, ... | | Authors: | Fujiwara, K, Hosaka, H, Nakagawa, A. | | Deposit date: | 2009-09-20 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Global conformational change associated with the two-step reaction catalyzed by Escherichia coli lipoate-protein ligase A.
J.Biol.Chem., 285, 2010
|
|
3A7U
 
 | |
7EXF
 
 | | Crystal structure of wild-type from Arabidopsis thaliana complexed with Galactose | | Descriptor: | Probable galactinol--sucrose galactosyltransferase 6, beta-D-galactopyranose | | Authors: | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-11-30 | | Last modified: | 2023-02-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7EXJ
 
 | | Crystal structure of alkaline alpha-galctosidase D383A mutant from Arabidopsis thaliana complexed with Raffinose | | Descriptor: | Probable galactinol--sucrose galactosyltransferase 6, alpha-D-galactopyranose-(1-6)-alpha-D-glucopyranose-(1-2)-beta-D-fructofuranose | | Authors: | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7EXQ
 
 | | Crystal structure of alkaline alpha-galactosidase D383A mutant from Arabidopsis thaliana complexed with product-galactose and sucrose. | | Descriptor: | Probable galactinol--sucrose galactosyltransferase 6, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, beta-D-galactopyranose | | Authors: | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | Deposit date: | 2021-05-28 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7EXR
 
 | | Crystal structure of alkaline alpha-galactosidase D383A mutant from Arabidopsis thaliana complexed with Stachyose. | | Descriptor: | Probable galactinol--sucrose galactosyltransferase 6, alpha-D-galactopyranose-(1-6)-alpha-D-galactopyranose-(1-6)-alpha-D-glucopyranose-(1-2)-beta-D-fructofuranose | | Authors: | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | Deposit date: | 2021-05-28 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7EXH
 
 | | Crystal structure of D383A mutant from Arabidopsis thaliana complexed with Galactinol. | | Descriptor: | Probable galactinol--sucrose galactosyltransferase 6, galactinol | | Authors: | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7EXG
 
 | | Crystal structure of D383A mutant from Arabidopsis thaliana complexed with Galactose. | | Descriptor: | Probable galactinol--sucrose galactosyltransferase 6, beta-D-galactopyranose | | Authors: | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
1ROM
 
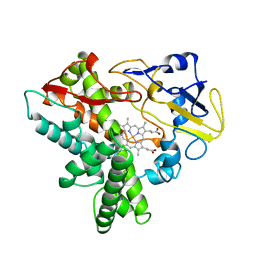 | |
1UKL
 
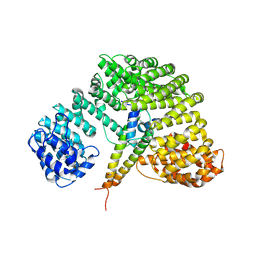 | | Crystal structure of Importin-beta and SREBP-2 complex | | Descriptor: | Importin beta-1 subunit, Sterol regulatory element binding protein-2 | | Authors: | Lee, S.J, Sekimoto, T, Yamashita, E, Nagoshi, E, Nakagawa, A, Imamoto, N, Yoshimura, M, Sakai, H, Tsukihara, T, Yoneda, Y. | | Deposit date: | 2003-08-26 | | Release date: | 2003-12-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of Importin-beta Bound to SREBP-2: Nuclear Import of a Transcription Factor
Science, 302, 2003
|
|
1UMK
 
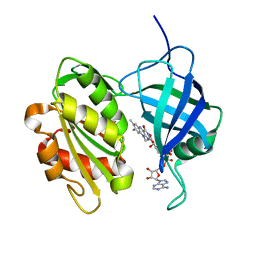 | | The Structure of Human Erythrocyte NADH-cytochrome b5 Reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase | | Authors: | Bando, S, Takano, T, Yubisui, T, Shirabe, K, Takeshita, M, Horii, C, Nakagawa, A. | | Deposit date: | 2003-10-03 | | Release date: | 2004-11-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of human erythrocyte NADH-cytochrome b5 reductase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3VMZ
 
 | |
3VRP
 
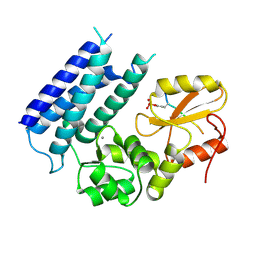 | | Crystal structure of the tyrosine kinase binding domain of Cbl-c in complex with phospho-EGFR peptide | | Descriptor: | CALCIUM ION, Epidermal growth factor receptor, Signal transduction protein CBL-C | | Authors: | Takeshita, K, Tezuka, T, Isozaki, Y, Yamashita, E, Suzuki, M, Yamanashi, Y, Yamamoto, T, Nakagawa, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of Cbl-c.
J.Biochem., 152, 2012
|
|
