7Z7X
 
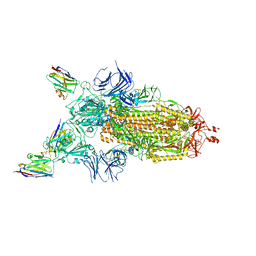 | | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11-H6 nanobody complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11-H6, ... | | Authors: | Weckener, M, Naismith, J.H. | | Deposit date: | 2022-03-16 | | Release date: | 2022-07-13 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
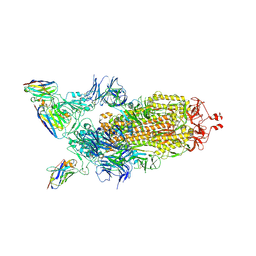
7Z86
 
 | |
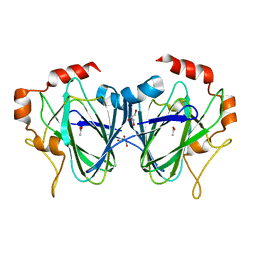
7Z6V
 
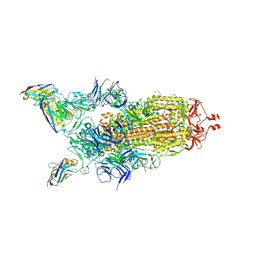 | | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11 nanobody complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11, ... | | Authors: | Weckener, M, Naismith, J.H, Vogirala, V.K. | | Deposit date: | 2022-03-14 | | Release date: | 2022-07-13 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6TSC
 
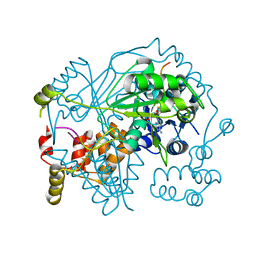 | | OphMA I407P complex with SAH | | Descriptor: | GLY-PHE-PRO-TRP-MVA-ILE-MVA-VAL-GLY-VAL-PRO-GLY, Peptide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2019-12-20 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Substrate Plasticity of a Fungal Peptide alpha-N-Methyltransferase.
Acs Chem.Biol., 15, 2020
|
|
1KCZ
 
 | | Crystal Structure of beta-methylaspartase from Clostridium tetanomorphum. Mg-complex. | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, beta-methylaspartase | | Authors: | Asuncion, M, Blankenfeldt, W, Barlow, J.N, Gani, D, Naismith, J.H. | | Deposit date: | 2001-11-12 | | Release date: | 2001-12-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of 3-methylaspartase from Clostridium tetanomorphum functions via the common enolase chemical step.
J.Biol.Chem., 277, 2002
|
|
1KD0
 
 | | Crystal Structure of beta-methylaspartase from Clostridium tetanomorphum. Apo-structure. | | Descriptor: | 1,2-ETHANEDIOL, beta-methylaspartase | | Authors: | Asuncion, M, Blankenfeldt, W, Barlow, J.N, Gani, D, Naismith, J.H. | | Deposit date: | 2001-11-12 | | Release date: | 2001-12-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of 3-methylaspartase from Clostridium tetanomorphum functions via the common enolase chemical step.
J.Biol.Chem., 277, 2002
|
|
1O7I
 
 | |
1OFN
 
 | | Purification, crystallisation and preliminary structural studies of dTDP-4-keto-6-deoxy-glucose-5-epimerase (EvaD) from Amycolatopsis orientalis; the fourth enzyme in the dTDP-L-epivancosamine biosynthetic pathway. | | Descriptor: | GLYCEROL, PCZA361.16 | | Authors: | Merkel, A.B, Naismith, J.H. | | Deposit date: | 2003-04-17 | | Release date: | 2004-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Purification, Crystallization and Preliminary Structural Studies of Dtdp-4-Keto-6-Deoxy-Glucose-5-Epimerase (Evad) from Amycolatopsis Orientalis, the Fourth Enzyme in the Dtdp-L-Epivancosamine Biosynthetic Pathway.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1PM7
 
 | |
1RTV
 
 | | RmlC (dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase) crystal structure from Pseudomonas aeruginosa, apo structure | | Descriptor: | S,R MESO-TARTARIC ACID, dTDP-4-dehydrorhamnose 3,5-epimerase | | Authors: | Dong, C.J, Naismith, J.H. | | Deposit date: | 2003-12-10 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RmlC (dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase) crystal structure from Pseudomonas aeruginosa, apo structure
TO BE PUBLISHED
|
|
7BKA
 
 | |
7BHJ
 
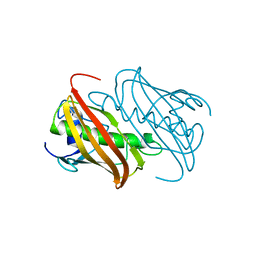 | |
6Z43
 
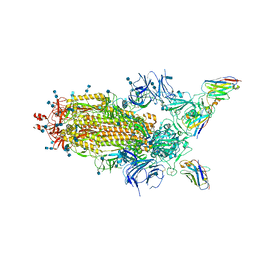 | | Cryo-EM Structure of SARS-CoV-2 Spike : H11-D4 Nanobody Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody, ... | | Authors: | Ruza, R.R, Duyvesteyn, H.M.E, Shah, P, Carrique, L, Ren, J, Malinauskas, T, Zhou, D, Stuart, D.I, Naismith, J.H. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for a potent neutralising single-domain antibody that blocks SARS-CoV-2 binding to its receptor ACE2
To Be Published
|
|
2IYC
 
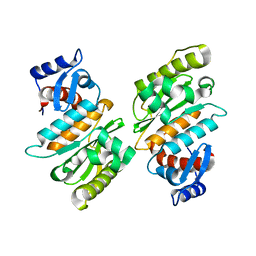 | | SENP1 native structure | | Descriptor: | SENTRIN-SPECIFIC PROTEASE 1 | | Authors: | Dong, C, Naismith, J.H. | | Deposit date: | 2006-07-14 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Senp1 Native Structure
To be Published
|
|
2IYD
 
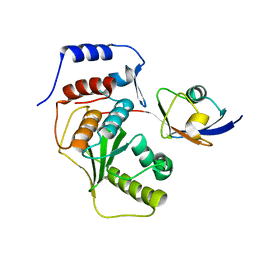 | | SENP1 covalent complex with SUMO-2 | | Descriptor: | SENTRIN-SPECIFIC PROTEASE 1, SMALL UBIQUITIN-RELATED MODIFIER 2 | | Authors: | Dong, C, Naismith, J.H. | | Deposit date: | 2006-07-14 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Senp1 Native Structure
To be Published
|
|
2JKC
 
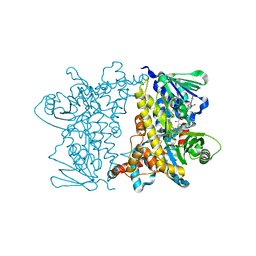 | | Crystal Structure of E346D of Tryptophan 7-Halogenase (PrnA) | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent tryptophan halogenase PrnA, ... | | Authors: | Zhu, X, Naismith, J.H. | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New insights into the mechanism of enzymatic chlorination of tryptophan.
Angew. Chem. Int. Ed. Engl., 47, 2008
|
|
6FU2
 
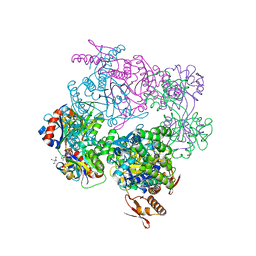 | | ATP phosphoribosyltransferase (HisZG ATPPRT) from Psychrobacter arcticus in complex with PRPP and ATP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Alphey, M.S, Ge, Y, Fisher, G, Czekster, C.M, Naismith, J.H, da Silva, R.G. | | Deposit date: | 2018-02-26 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Catalytic and Anticatalytic Snapshots of a Short-Form ATP Phosphoribosyltransferase
Acs Catalysis, 2018
|
|
6FU7
 
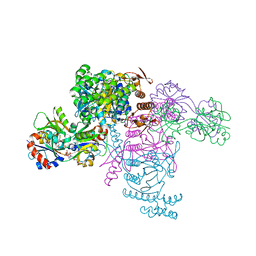 | | ATP phosphoribosyltransferase (HisZG ATPPRT) from Psychrobacter arcticus in complex with PRATP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ATP phosphoribosyltransferase, ATP phosphoribosyltransferase regulatory subunit, ... | | Authors: | Alphey, M.S, Ge, Y, Fisher, G, Czekster, C.M, Naismith, J.H, da Silva, R.G. | | Deposit date: | 2018-02-26 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Catalytic and Anticatalytic Snapshots of a Short-Form ATP Phosphoribosyltransferase
Acs Catalysis, 2018
|
|
6GI2
 
 | |
6GI5
 
 | | Crystal structure of the ferric enterobactin esterase (PfeE) from Pseudomonas aeruginosa in complex with the tris-catechol vector | | Descriptor: | FE (III) ION, Ferric enterobactin esterase, ~{N}-[2-[[(2~{S})-2-[[2,3-bis(oxidanyl)phenyl]carbonylamino]-3-[[(2~{S})-2-[[2,3-bis(oxidanyl)phenyl]carbonylamino]-3-oxidanylidene-3-(prop-2-ynylamino)propyl]amino]-3-oxidanylidene-propyl]amino]-2-oxidanylidene-ethyl]-2,3-bis(oxidanyl)benzamide | | Authors: | Moynie, L, Naismith, J.H. | | Deposit date: | 2018-05-09 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | A Key Role for the Periplasmic PfeE Esterase in Iron Acquisition via the Siderophore Enterobactin in Pseudomonas aeruginosa.
ACS Chem. Biol., 13, 2018
|
|
6GI1
 
 | | Crystal structure of the ferric enterobactin esterase (pfeE) mutant(S157A) from Pseudomonas aeruginosa in presence of enterobactin | | Descriptor: | 1,2-ETHANEDIOL, 2-(2,3-DIHYDROXY-BENZOYLAMINO)-3-HYDROXY-PROPIONIC ACID, FE (III) ION, ... | | Authors: | Moynie, L, Naismith, J.H. | | Deposit date: | 2018-05-09 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | A Key Role for the Periplasmic PfeE Esterase in Iron Acquisition via the Siderophore Enterobactin in Pseudomonas aeruginosa.
ACS Chem. Biol., 13, 2018
|
|
6GEW
 
 | | OphA Y63F-sinefungin complex | | Descriptor: | OphA, S-ADENOSYL-L-HOMOCYSTEINE, SINEFUNGIN | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2018-04-27 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A molecular mechanism for the enzymatic methylation of nitrogen atoms within peptide bonds.
Sci Adv, 4, 2018
|
|
6GI0
 
 | |
6H7F
 
 | | Crystal structure of BauA, the Ferric preacinetobactin receptor from Acinetobacter baumannii in complex with Fe3+-Preacinetobactin-acinetobactin | | Descriptor: | (4~{S},5~{R})-2-[2,3-bis(oxidanyl)phenyl]-~{N}-[2-(1~{H}-imidazol-4-yl)ethyl]-5-methyl-~{N}-oxidanyl-4,5-dihydro-1,3-oxazole-4-carboxamide, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-ETHANEDIOL, ... | | Authors: | Moynie, L, Naismith, J.H. | | Deposit date: | 2018-07-31 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Preacinetobactin not acinetobactin is essential for iron uptake by the BauA transporter of the pathogenAcinetobacter baumannii.
Elife, 7, 2018
|
|
6H7V
 
 | |
