5N0R
 
 | |
5N4I
 
 | |
5NC4
 
 | | Crystal structure of the ferric enterobactin receptor (PfeA) in complex with protochelin from Pseudomonas aeruginosa | | 分子名称: | FE (III) ION, Ferric enterobactin receptor, ~{N}-[(5~{S})-5-[[2,3-bis(oxidanyl)phenyl]carbonylamino]-6-[4-[[2,3-bis(oxidanyl)phenyl]carbonylamino]butylamino]-6-oxidanylidene-hexyl]-2,3-bis(oxidanyl)benzamide | | 著者 | Moynie, L, Naismith, J.H. | | 登録日 | 2017-03-03 | | 公開日 | 2018-03-21 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The complex of ferric-enterobactin with its transporter from Pseudomonas aeruginosa suggests a two-site model.
Nat Commun, 10, 2019
|
|
5O3X
 
 | | Structural characterization of the fast and promiscuous macrocyclase from plant - apo PCY1 | | 分子名称: | CACODYLATE ION, Peptide cyclase 1 | | 著者 | Ludewig, H, Czekster, C.M, Bent, A.F, Naismith, J.H. | | 登録日 | 2017-05-25 | | 公開日 | 2018-02-07 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Characterization of the Fast and Promiscuous Macrocyclase from Plant PCY1 Enables the Use of Simple Substrates.
ACS Chem. Biol., 13, 2018
|
|
5O3V
 
 | | Structural characterization of the fast and promiscuous macrocyclase from plant - PCY1-S562A bound to Presegetalin B1 | | 分子名称: | MAGNESIUM ION, Peptide cyclase 1, Putative presegetalin B1, ... | | 著者 | Ludewig, H, Czekster, C.M, Bent, A.F, Naismith, J.H. | | 登録日 | 2017-05-25 | | 公開日 | 2018-02-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Characterization of the Fast and Promiscuous Macrocyclase from Plant PCY1 Enables the Use of Simple Substrates.
ACS Chem. Biol., 13, 2018
|
|
1WA3
 
 | | Mechanism of the Class I KDPG aldolase | | 分子名称: | 2-KETO-3-DEOXY-6-PHOSPHOGLUCONATE ALDOLASE, PYRUVIC ACID, SULFATE ION | | 著者 | Fullerton, S.W.B, Griffiths, J.S, Merkel, A.B, Wymer, N.J, Hutchins, M.J, Fierke, C.A, Toone, E.J, Naismith, J.H. | | 登録日 | 2004-10-22 | | 公開日 | 2005-01-26 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Mechanism of the Class I Kdpg Aldolase.
Bioorg.Med.Chem., 14, 2006
|
|
5O3U
 
 | | Structural characterization of the fast and promiscuous macrocyclase from plant - PCY1-S562A bound to Presegetalin F1 | | 分子名称: | Peptide cyclase 1, Putative presegetalin F1 | | 著者 | Ludewig, H, Czekster, C.M, Bent, A.F, Naismith, J.H. | | 登録日 | 2017-05-25 | | 公開日 | 2018-02-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Characterization of the Fast and Promiscuous Macrocyclase from Plant PCY1 Enables the Use of Simple Substrates.
ACS Chem. Biol., 13, 2018
|
|
1WAU
 
 | |
1WBH
 
 | |
1WAM
 
 | |
5O3W
 
 | | Structural characterization of the fast and promiscuous macrocyclase from plant - PCY1-S562A bound to Presegetalin A1 | | 分子名称: | MAGNESIUM ION, Peptide cyclase 1, Presegetalin A1, ... | | 著者 | Ludewig, H, Czekster, C.M, Bent, A.F, Naismith, J.H. | | 登録日 | 2017-05-25 | | 公開日 | 2018-02-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Characterization of the Fast and Promiscuous Macrocyclase from Plant PCY1 Enables the Use of Simple Substrates.
ACS Chem. Biol., 13, 2018
|
|
5OUF
 
 | |
5OUT
 
 | |
7Z1A
 
 | | Nanobody H11 and F2 bound to RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F2 Nanobody, H11 Nanobody, ... | | 著者 | Mikolajek, H, Naismith, J.H. | | 登録日 | 2022-02-24 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z1D
 
 | | Nanobody H11-H6 bound to RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, H11-H6 nanobody, ... | | 著者 | Mikolajek, H, Naismith, J.H. | | 登録日 | 2022-02-24 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z1E
 
 | | Nanobody H11-H4 Q98R H100E bound to RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, H11-H4 Q98R H100E, ... | | 著者 | Mikolajek, H, Naismith, J.H. | | 登録日 | 2022-02-24 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z1C
 
 | | Nanobody H11-B5 and H11-F2 bound to RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Nanobody B5, ... | | 著者 | Mikolajek, H, Naismith, J.H. | | 登録日 | 2022-02-24 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z1B
 
 | | Nanobody H11-A10 and F2 bound to RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody A10, Nanobody F2, ... | | 著者 | Mikolajek, H, Naismith, J.H. | | 登録日 | 2022-02-24 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZB0
 
 | | macrocyclase OphP with 15mer | | 分子名称: | 1,2-ETHANEDIOL, 15mer, BICARBONATE ION, ... | | 著者 | Song, H, Naismith, J.H. | | 登録日 | 2022-03-23 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Molecular basis for the enzymatic macrocyclization of multiply backbone N-methylated peptides
Biorxiv, 2022
|
|
7ZB1
 
 | | S580A with 18mer | | 分子名称: | 1,2-ETHANEDIOL, 18mer, BICARBONATE ION, ... | | 著者 | Song, H, Naismith, J.H. | | 登録日 | 2022-03-23 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Molecular basis for the enzymatic macrocyclization of multiply backbone N-methylated peptides
Biorxiv, 2022
|
|
7ZAZ
 
 | | macrocyclase OphP with ZPP | | 分子名称: | 1,2-ETHANEDIOL, BICARBONATE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Song, H, Naismith, J.H. | | 登録日 | 2022-03-23 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Molecular basis for the enzymatic macrocyclization of multiply backbone N-methylated peptides
Biorxiv, 2022
|
|
7ZB2
 
 | | apo macrocyclase OphP | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Song, H, Naismith, J.H. | | 登録日 | 2022-03-23 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Molecular basis for the enzymatic macrocyclization of multiply backbone N-methylated peptides
Biorxiv, 2022
|
|
7Z9R
 
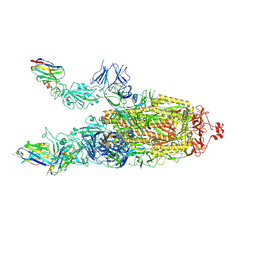 | |
7Z9Q
 
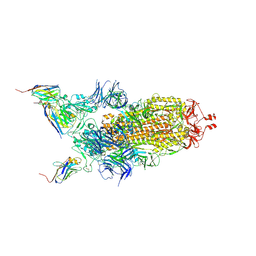 | | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11-A10 nanobody complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11-A10, ... | | 著者 | Weckener, M, Naismith, J.H. | | 登録日 | 2022-03-21 | | 公開日 | 2022-07-13 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z85
 
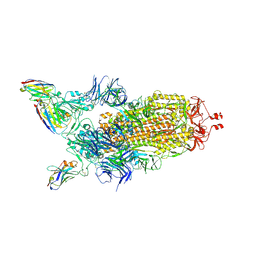 | | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11-B5 nanobody complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11-B5, ... | | 著者 | Weckener, M, Naismith, J.H. | | 登録日 | 2022-03-16 | | 公開日 | 2022-07-13 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
