3KYX
 
 | |
7TX3
 
 | |
5TC9
 
 | |
5TKI
 
 | |
5TKH
 
 | |
5TKF
 
 | | Neurospora crassa polysaccharide monooxygenase 2 high mannosylation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | 著者 | O'Dell, W.B, Meilleur, F. | | 登録日 | 2016-10-06 | | 公開日 | 2017-05-17 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystallization of a fungal lytic polysaccharide monooxygenase expressed from glycoengineered Pichia pastoris for X-ray and neutron diffraction.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5TKG
 
 | | Neurospora crassa polysaccharide monooxygenase 2 resting state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Lytic polysaccharide monooxygenase, ... | | 著者 | O'Dell, W.B, Meilleur, F. | | 登録日 | 2016-10-06 | | 公開日 | 2017-05-17 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Oxygen Activation at the Active Site of a Fungal Lytic Polysaccharide Monooxygenase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5VNQ
 
 | | Neutron crystallographic structure of perdeuterated T4 lysozyme cysteine-free pseudo-wild type at cryogenic temperature | | 分子名称: | CHLORIDE ION, Endolysin | | 著者 | Li, L, Shukla, S, Meilleur, F, Standaert, R.F, Pierce, J, Myles, D.A.A, Cuneo, M.J. | | 登録日 | 2017-05-01 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-10-04 | | 実験手法 | NEUTRON DIFFRACTION (2.2 Å) | | 主引用文献 | Neutron crystallographic studies of T4 lysozyme at cryogenic temperature.
Protein Sci., 26, 2017
|
|
5VNR
 
 | | X-ray structure of perdeuterated T4 lysozyme cysteine-free pseudo-wild type at cryogenic temperature | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Endolysin, ... | | 著者 | Li, L, Shukla, S, Meilleur, F, Standaert, R.F, Pierce, J, Myles, D.A.A, Cuneo, M.J. | | 登録日 | 2017-05-01 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.631 Å) | | 主引用文献 | Neutron crystallographic studies of T4 lysozyme at cryogenic temperature.
Protein Sci., 26, 2017
|
|
7PXR
 
 | | Room temperature structure of an LPMO. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, CHLORIDE ION, ... | | 著者 | Tandrup, T, Meilleur, F, Ipsen, J, Johansen, K.S, Lo Leggio, L. | | 登録日 | 2021-10-08 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
6MEZ
 
 | | X-ray structure of the Fenna-Matthews-Olsen antenna complex from Prosthecochloris aestuarii | | 分子名称: | BACTERIOCHLOROPHYLL A, Bacteriochlorophyll a protein, SULFATE ION | | 著者 | Selvaraj, B, Lu, X, Cuneo, M.J, Myles, D.A.A. | | 登録日 | 2018-09-07 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Neutron and X-ray analysis of the Fenna-Matthews-Olson photosynthetic antenna complex from Prosthecochloris aestuarii.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
4XWR
 
 | |
4XXG
 
 | |
5EAJ
 
 | |
7ADQ
 
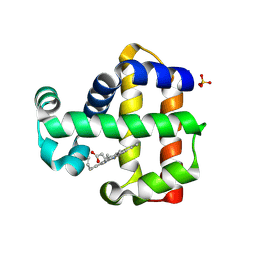 | | Serial Laue crystallography structure of dehaloperoxidase B from Amphitrite ornata | | 分子名称: | Dehaloperoxidase B, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | 著者 | Moreno-Chicano, T.M, Ebrahim, A.E, Srajer, V, Henning, R.W, Doak, B.C, Trebbin, M, Monteiro, D.C.F, Hough, M.A. | | 登録日 | 2020-09-15 | | 公開日 | 2021-11-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Complementarity of neutron, XFEL and synchrotron crystallography for defining the structures of metalloenzymes at room temperature.
Iucrj, 9, 2022
|
|
5XPE
 
 | | Neutron structure of the T26H mutant of T4 lysozyme | | 分子名称: | CHLORIDE ION, Endolysin, SODIUM ION | | 著者 | Hiromoto, T, Kuroki, R. | | 登録日 | 2017-06-01 | | 公開日 | 2017-10-04 | | 最終更新日 | 2024-04-03 | | 実験手法 | NEUTRON DIFFRACTION (1.648 Å), X-RAY DIFFRACTION | | 主引用文献 | Neutron structure of the T26H mutant of T4 phage lysozyme provides insight into the catalytic activity of the mutant enzyme and how it differs from that of wild type.
Protein Sci., 26, 2017
|
|
5XPF
 
 | |
7KFM
 
 | | Room temperature oxyferrous Dehaloperoxidase B | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, Dehaloperoxidase B, OXYGEN MOLECULE, ... | | 著者 | Carey, L.M, Ghiladi, R.A. | | 登録日 | 2020-10-14 | | 公開日 | 2021-10-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Complementarity of neutron, XFEL and synchrotron crystallography for defining the structures of metalloenzymes at room temperature.
Iucrj, 9, 2022
|
|
3KYU
 
 | |
3KYV
 
 | |
3KYW
 
 | |
3KYY
 
 | |
7TOB
 
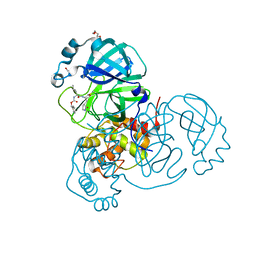 | | Crystal structure of the SARS-CoV-2 Omicron main protease (Mpro) in complex with inhibitor GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | 著者 | Sacco, M.D, Wang, J, Chen, Y. | | 登録日 | 2022-01-24 | | 公開日 | 2022-02-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | The P132H mutation in the main protease of Omicron SARS-CoV-2 decreases thermal stability without compromising catalysis or small-molecule drug inhibition.
Cell Res., 32, 2022
|
|
7TWS
 
 | |
7TWI
 
 | |
