3ME8
 
 | | Crystal structure of putative electron transfer protein aq_2194 from Aquifex aeolicus VF5 | | Descriptor: | Putative uncharacterized protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-14 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of putative electron transfer protein aq_2194 from Aquifex aeolicus VF5
To be Published
|
|
3MF4
 
 | | Crystal structure of putative two-component system response regulator/ggdef domain protein | | Descriptor: | MAGNESIUM ION, Two-component system response regulator/GGDEF domain protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-01 | | Release date: | 2010-04-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of putative two-component system response regulator/ggdef domain protein
To be Published
|
|
3M0G
 
 | | CRYSTAL STRUCTURE OF putative farnesyl diphosphate synthase from Rhodobacter capsulatus | | Descriptor: | Farnesyl diphosphate synthase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-03 | | Release date: | 2010-03-31 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CRYSTAL STRUCTURE OF putative farnesyl diphosphate synthase from Rhodobacter capsulatus
To be Published
|
|
3N4E
 
 | | CRYSTAL STRUCTURE OF mandelate racemase/muconate lactonizing protein from Paracoccus denitrificans Pd1222 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Mandelate racemase/muconate lactonizing enzyme, ... | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CRYSTAL STRUCTURE OF mandelate racemase/muconate lactonizing protein from Paracoccus denitrificans
Pd1222
To be Published
|
|
3N4F
 
 | | CRYSTAL STRUCTURE OF Mandelate racemase/muconate lactonizing protein from Geobacillus sp. Y412MC10 | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | CRYSTAL STRUCTURE OF Mandelate racemase/muconate lactonizing protein from Geobacillus
sp. Y412MC10
To be Published
|
|
3N6J
 
 | | Crystal structure of Mandelate racemase/muconate lactonizing protein from Actinobacillus succinogenes 130Z | | Descriptor: | Mandelate racemase/muconate lactonizing protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-25 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Mandelate racemase/muconate lactonizing protein from Actinobacillus
succinogenes 130Z
To be Published
|
|
3N6H
 
 | | Crystal structure of Mandelate racemase/muconate lactonizing protein from Actinobacillus succinogenes 130Z complexed with magnesium/sulfate | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein, ... | | Authors: | Malashkevich, V.N, Patskovsky, Y, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-25 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Mandelate racemase/muconate lactonizing protein from Actinobacillus
succinogenes 130Z complexed with magnesium/sulfate
To be Published
|
|
3OA4
 
 | | CRYSTAL STRUCTURE OF hypothetical protein BH1468 from Bacillus halodurans C-125 | | Descriptor: | GLYCEROL, SULFATE ION, ZINC ION, ... | | Authors: | Malashkevich, V.N, Patskovsky, Y, Garrett, S, Foti, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-08-04 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | CRYSTAL STRUCTURE OF hypothetical protein BH1468 from Bacillus halodurans C-125
To be Published
|
|
3OAN
 
 | | Crystal structure of the Ran Binding Domain From The Nuclear Complex Component Nup2 From Ashbya Gossypii | | Descriptor: | ABR034Wp, GLYCEROL | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-08-05 | | Release date: | 2010-08-18 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Ran Binding Domain From The
Nuclear Complex Component Nup2 From Ashbya Gossypii
To be Published
|
|
3OAJ
 
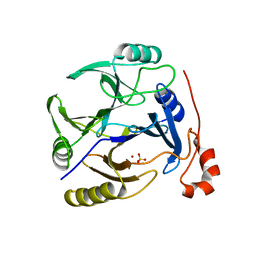 | | CRYSTAL STRUCTURE OF putative dioxygenase from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | Putative ring-cleaving dioxygenase mhqO, SULFATE ION, ZINC ION | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-08-05 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | CRYSTAL STRUCTURE OF putative dioxygenase from Bacillus subtilis subsp. subtilis str.
168
To be Published
|
|
3OP2
 
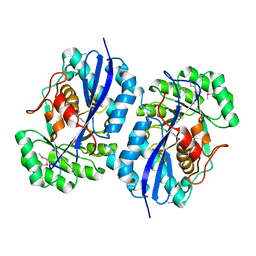 | | Crystal structure of putative mandelate racemase from Bordetella bronchiseptica RB50 complexed with 2-oxoglutarate/phosphate | | Descriptor: | 2-OXOGLUTARIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Malashkevich, V.N, Patskovsky, Y, Ramagopal, U, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of putative mandelate racemase from
Bordetella bronchiseptica RB50 complexed with 2-oxoglutarate/phosphate
To be Published
|
|
3OPS
 
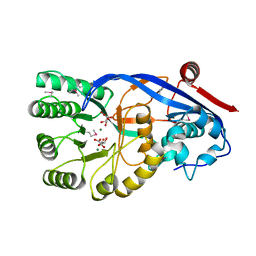 | | Crystal structure of mandelate racemase/muconate lactonizing protein FROM GEOBACILLUS SP. Y412MC10 complexed with magnesium/tartrate | | Descriptor: | D(-)-TARTARIC ACID, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Malashkevich, V.N, Patskovsky, Y, Ramagopal, U, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-09-01 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure ofmandelate racemase/muconate lactonizing protein FROM GEOBACILLUS SP. Y412MC10 complexed with magnesium/tartrate
To be Published
|
|
3OOQ
 
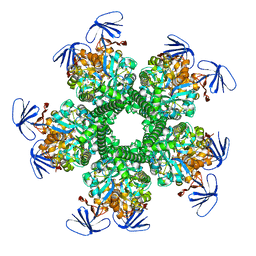 | | CRYSTAL STRUCTURE OF amidohydrolase from Thermotoga maritima MSB8 | | Descriptor: | GLYCEROL, amidohydrolase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-15 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | CRYSTAL STRUCTURE OF amidohydrolase from Thermotoga maritima MSB8
To be Published
|
|
3OQB
 
 | | CRYSTAL STRUCTURE OF putative oxidoreductase from Bradyrhizobium japonicum USDA 110 | | Descriptor: | Oxidoreductase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-09-02 | | Release date: | 2010-09-15 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CRYSTAL STRUCTURE OF putative oxidoreductase from Bradyrhizobium japonicum USDA 110
To be Published
|
|
3PQA
 
 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase GapN from Methanocaldococcus jannaschii DSM 2661 | | Descriptor: | Lactaldehyde dehydrogenase, SULFATE ION | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-11-25 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase GapN from Methanocaldococcus jannaschii DSM 2661
To be Published
|
|
3PTW
 
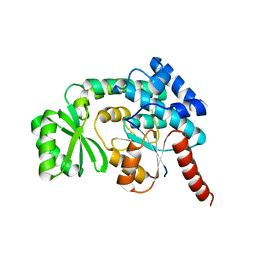 | | CRYSTAL STRUCTURE OF malonyl CoA-acyl carrier protein transacylase from Clostridium perfringens Atcc 13124 | | Descriptor: | Malonyl CoA-acyl carrier protein transacylase | | Authors: | Malashkevich, V.N, Toro, R, Ramagopal, U, Seidel, R, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-12-03 | | Release date: | 2010-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CRYSTAL STRUCTURE OF malonyl CoA-acyl carrier protein transacylase from Clostridium perfringens
Atcc 13124
To be Published
|
|
3PRL
 
 | | Crystal structure of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Bacillus halodurans C-125 | | Descriptor: | NADP-dependent glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-11-29 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Bacillus halodurans C-125
To be Published
|
|
3QKC
 
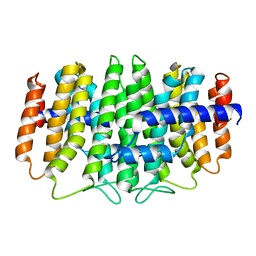 | | CRYSTAL STRUCTURE OF geranyl diphosphate synthase small subunit from Antirrhinum majus | | Descriptor: | Geranyl diphosphate synthase small subunit | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of geranyl diphosphate synthase small subunit from Antirrhinum majus
To be Published
|
|
3QK7
 
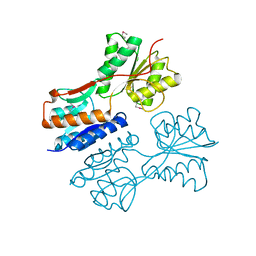 | | Crystal structure of putative Transcriptional regulator from Yersinia pestis biovar Microtus str. 91001 | | Descriptor: | Transcriptional regulators | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of putative Transcriptional regulator from Yersinia pestis biovar Microtus str. 91001
To be Published
|
|
3RMT
 
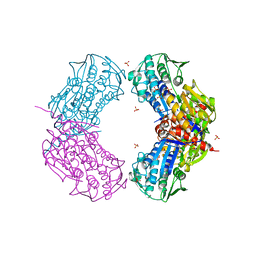 | | Crystal structure of putative 5-enolpyruvoylshikimate-3-phosphate synthase from Bacillus halodurans C-125 | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase 1, SULFATE ION | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Ramagopal, U, Zencheck, W, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-04-21 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of putative 5-enolpyruvoylshikimate-3-phosphate synthase from Bacillus halodurans C-125
To be Published
|
|
3RCY
 
 | | CRYSTAL STRUCTURE OF Mandelate racemase/muconate lactonizing enzyme-like protein from Roseovarius sp. TM1035 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme-like protein, ... | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-03-31 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | CRYSTAL STRUCTURE OF Mandelate racemase/muconate lactonizing enzyme-like protein from Roseovarius sp. TM1035
To be Published
|
|
3RHD
 
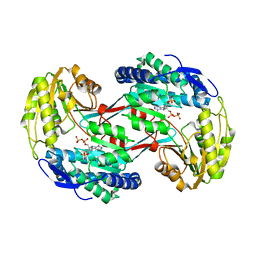 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase GapN from Methanocaldococcus jannaschii DSM 2661 complexed with NADP | | Descriptor: | Lactaldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-04-11 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase GapN from Methanocaldococcus jannaschii DSM 2661 complexed with NADP
To be Published
|
|
4K8P
 
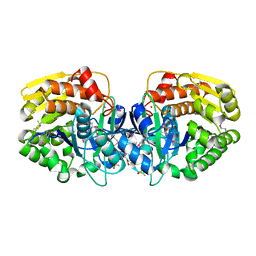 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 2-ethylbenzyl alcohol | | Descriptor: | (2-ETHYLPHENYL)METHANOL, ADENOSINE, DIMETHYL SULFOXIDE, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 2-ethylbenzyl alcohol
To be Published
|
|
4KAN
 
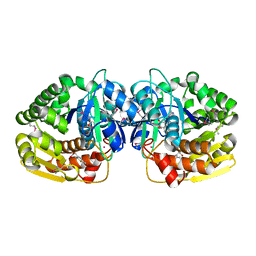 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 2-(2,5-dimethyl-1,3-thiazol-4-yl)acetic acid | | Descriptor: | (2,5-dimethyl-1,3-thiazol-4-yl)acetic acid, ADENOSINE, POTASSIUM ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-22 | | Release date: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 2-(2,5-dimethyl-1,3-thiazol-4-yl)acetic acid
To be Published
|
|
4KAD
 
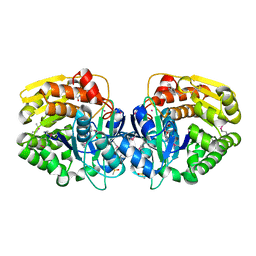 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with N1-(2.3-dihydro-1H-inden-5-yl)acetam | | Descriptor: | ADENOSINE, DIMETHYL SULFOXIDE, N-(4,7-dihydro-1H-inden-6-yl)acetamide, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-22 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with N1-(2.3-dihydro-1H-inden-5-yl)acetam
To be Published
|
|
