3WJO
 
 | | Crystal structure of Octaprenyl Pyrophosphate synthase from Escherichia coli with isopentenyl pyrophosphate (IPP) | | 分子名称: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Octaprenyl diphosphate synthase | | 著者 | Han, X, Chen, C.C, Kuo, C.J, Huang, C.H, Zheng, Y, Ko, T.P, Zhu, Z, Feng, X, Oldfield, E, Liang, P.H, Guo, R.T, Ma, Y.H. | | 登録日 | 2013-10-12 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Crystal structures of ligand-bound octaprenyl pyrophosphate synthase from Escherichia coli reveal the catalytic and chain-length determining mechanisms.
Proteins, 83, 2015
|
|
3WJK
 
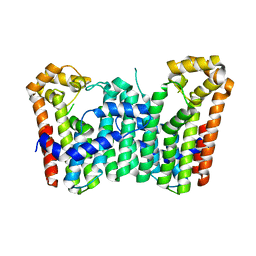 | | Crystal structure of Octaprenyl Pyrophosphate synthase from Escherichia coli | | 分子名称: | Octaprenyl diphosphate synthase | | 著者 | Han, X, Chen, C.C, Kuo, C.J, Huang, C.H, Zheng, Y, Ko, T.P, Zhu, Z, Feng, X, Oldfield, E, Liang, P.H, Guo, R.T, Ma, Y.H. | | 登録日 | 2013-10-11 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structures of ligand-bound octaprenyl pyrophosphate synthase from Escherichia coli reveal the catalytic and chain-length determining mechanisms.
Proteins, 83, 2015
|
|
3WJN
 
 | | Crystal structure of Octaprenyl Pyrophosphate synthase from Escherichia coli with farnesyl S-thiol-pyrophosphate (FSPP) | | 分子名称: | Octaprenyl diphosphate synthase, S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE | | 著者 | Han, X, Chen, C.C, Kuo, C.J, Huang, C.H, Zheng, Y, Ko, T.P, Zhu, Z, Feng, X, Oldfield, E, Liang, P.H, Guo, R.T, Ma, Y.H. | | 登録日 | 2013-10-12 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structures of ligand-bound octaprenyl pyrophosphate synthase from Escherichia coli reveal the catalytic and chain-length determining mechanisms.
Proteins, 83, 2015
|
|
5TBP
 
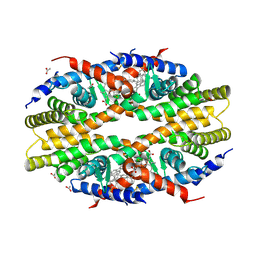 | | Crystal Structure of RXR-alpha ligand binding domain complexed with synthetic modulator K8003 | | 分子名称: | ACETATE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Aleshin, A.E, Liddington, R.C, Su, Y, Zhang, X. | | 登録日 | 2016-09-12 | | 公開日 | 2017-08-09 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Modulation of nongenomic activation of PI3K signalling by tetramerization of N-terminally-cleaved RXR alpha.
Nat Commun, 8, 2017
|
|
3VSV
 
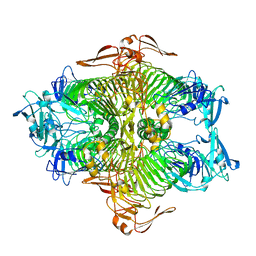 | | The complex structure of XylC with xylose | | 分子名称: | Xylosidase, alpha-D-xylopyranose, beta-D-xylopyranose | | 著者 | Huang, C.H, Sun, Y, Ko, T.P, Ma, Y, Chen, C.C, Zheng, Y, Chan, H.C, Pang, X, Wiegel, J, Shao, W, Guo, R.T. | | 登録日 | 2012-05-09 | | 公開日 | 2013-02-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | The substrate/product-binding modes of a novel GH120 beta-xylosidase (XylC) from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Biochem.J., 448, 2012
|
|
5GT2
 
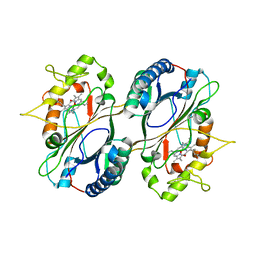 | | Crystal Structure and Biochemical Features of dye-decolorizing peroxidase YfeX from Escherichia coli O157 | | 分子名称: | PROTOPORPHYRIN IX CONTAINING FE, Probable deferrochelatase/peroxidase YfeX | | 著者 | Ma, Y.L, Yuan, Z.G, Liu, S, Wang, J.X, Gu, L.C, Liu, X.H. | | 登録日 | 2016-08-18 | | 公開日 | 2017-02-08 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.093 Å) | | 主引用文献 | Crystal structure and biochemical features of dye-decolorizing peroxidase YfeX from Escherichia coli O157 Asp(143) and Arg(232) play divergent roles toward different substrates
Biochem. Biophys. Res. Commun., 484, 2017
|
|
7E7X
 
 | |
7E7Y
 
 | |
7E8C
 
 | | SARS-CoV-2 S-6P in complex with 9 Fabs | | 分子名称: | 368-2 H, 368-2 L, 604 H, ... | | 著者 | Du, S, Xiao, J, Zhang, Z. | | 登録日 | 2021-03-01 | | 公開日 | 2021-06-09 | | 最終更新日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E88
 
 | |
7E8F
 
 | | SARS-CoV-2 NTD in complex with N9 Fab | | 分子名称: | 368-2 H, 368-2 L, 604 H, ... | | 著者 | Du, S, Xiao, J, Zhang, Z. | | 登録日 | 2021-03-01 | | 公開日 | 2021-06-09 | | 最終更新日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E86
 
 | |
3NCX
 
 | |
3NCW
 
 | |
7E7D
 
 | |
3VNI
 
 | | Crystal structures of D-Psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars | | 分子名称: | MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | 著者 | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | 登録日 | 2012-01-16 | | 公開日 | 2012-08-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
3VNJ
 
 | | Crystal structures of D-Psicose 3-epimerase with D-psicose from Clostridium cellulolyticum H10 | | 分子名称: | D-psicose, MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | 著者 | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | 登録日 | 2012-01-16 | | 公開日 | 2012-08-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
3VNM
 
 | | Crystal structures of D-Psicose 3-epimerase with D-sorbose from Clostridium cellulolyticum H10 | | 分子名称: | D-sorbose, MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | 著者 | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | 登録日 | 2012-01-17 | | 公開日 | 2012-08-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
3VNK
 
 | | Crystal structures of D-Psicose 3-epimerase with D-fructose from Clostridium cellulolyticum H10 | | 分子名称: | D-fructose, MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | 著者 | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | 登録日 | 2012-01-16 | | 公開日 | 2012-08-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
3VNL
 
 | | Crystal structures of D-Psicose 3-epimerase with D-tagatose from Clostridium cellulolyticum H10 | | 分子名称: | D-tagatose, MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | 著者 | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | 登録日 | 2012-01-16 | | 公開日 | 2012-08-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
7E7B
 
 | | Cryo-EM structure of the SARS-CoV-2 furin site mutant S-Trimer from a subunit vaccine candidate | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-hydroxyethyl 2-deoxy-3,5-bis-O-(2-hydroxyethyl)-6-O-(2-{[(9E)-octadec-9-enoyl]oxy}ethyl)-alpha-L-xylo-hexofuranoside, ... | | 著者 | Zheng, S, Ma, J. | | 登録日 | 2021-02-25 | | 公開日 | 2021-03-24 | | 最終更新日 | 2022-12-07 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Cryo-EM structure of S-Trimer, a subunit vaccine candidate for COVID-19.
J.Virol., 95, 2021
|
|
5WMG
 
 | | N-terminal bromodomain of BRD4 in complex with OTX-015 | | 分子名称: | 1,2-ETHANEDIOL, 4-{6-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1S)-1-(pyridin-2-yl)ethyl]-1H-pyrrolo[3,2-b]pyridin-3-yl}benzoic acid, Bromodomain-containing protein 4 | | 著者 | Zhang, Y. | | 登録日 | 2017-07-28 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.19 Å) | | 主引用文献 | BRD4 Profiling Identifies Critical Chronic Lymphocytic Leukemia Oncogenic Circuits and Reveals Sensitivity to PLX51107, a Novel Structurally Distinct BET Inhibitor.
Cancer Discov, 8, 2018
|
|
5WMD
 
 | | N-terminal bromodomain of BRD4 in complex with OTX-015 | | 分子名称: | 2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]-N-(4-hydroxyphenyl)acetamide, Bromodomain-containing protein 4 | | 著者 | Zhang, Y. | | 登録日 | 2017-07-28 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | BRD4 Profiling Identifies Critical Chronic Lymphocytic Leukemia Oncogenic Circuits and Reveals Sensitivity to PLX51107, a Novel Structurally Distinct BET Inhibitor.
Cancer Discov, 8, 2018
|
|
5WMA
 
 | | N-terminal bromodomain of BRD4 in complex with PLX5981 | | 分子名称: | 1,2-ETHANEDIOL, 5-(3,5-dimethyl-1,2-oxazol-4-yl)-1H-pyrrolo[2,3-b]pyridine, Bromodomain-containing protein 4 | | 著者 | Zhang, Y. | | 登録日 | 2017-07-28 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.401 Å) | | 主引用文献 | BRD4 Profiling Identifies Critical Chronic Lymphocytic Leukemia Oncogenic Circuits and Reveals Sensitivity to PLX51107, a Novel Structurally Distinct BET Inhibitor.
Cancer Discov, 8, 2018
|
|
7RPI
 
 | | Cryo-EM structure of murine Dispatched 'T' conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Lauryl Maltose Neopentyl Glycol, ... | | 著者 | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | 登録日 | 2021-08-03 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
