6JZ6
 
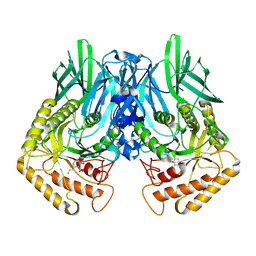 | | b-glucuronidase from Ruminococcus gnavus in complex with C6-substituted uronic isofagomine | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-4,5-bis(oxidanyl)-2-propyl-piperidine-3-carboxylic acid, Beta-glucuronidase | | Authors: | Dashnyam, P, Lin, H.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.605 Å) | | Cite: | Substituent Position of Iminocyclitols Determines the Potency and Selectivity for Gut Microbial Xenobiotic-Reactivating Enzymes.
J.Med.Chem., 63, 2020
|
|
3EX8
 
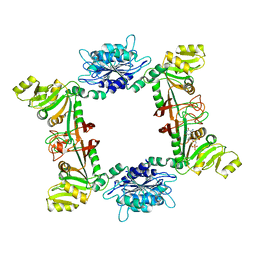 | | Complex structure of bacillus subtilis RibG reduction mechanism in riboflavin biosynthesis | | Descriptor: | Riboflavin biosynthesis protein ribD, ZINC ION, [(2R,3S,4R,5E)-5-[(5-amino-2,6-dioxo-3H-pyrimidin-4-yl)imino]-2,3,4-trihydroxy-pentyl] dihydrogen phosphate | | Authors: | Chen, S.C, Lin, Y.H, Yu, H.C, Liaw, S.H. | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Complex structure of Bacillus subtilis RibG: the reduction mechanism during riboflavin biosynthesis.
J.Biol.Chem., 284, 2009
|
|
7ZO1
 
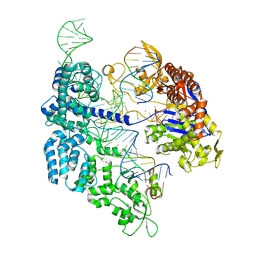 | | SpCas9 bound to CD34 off-target9 DNA substrate | | Descriptor: | CD34 off-target9 DNA non-target strand, CD34 off-target9 DNA target strand, CD34 sgRNA, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-04-23 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
3NOA
 
 | |
3SIX
 
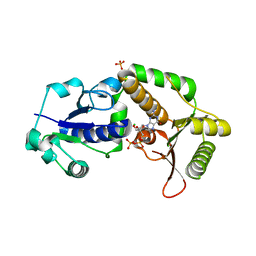 | | Crystal structure of NodZ alpha-1,6-fucosyltransferase soaked with GDP-fucose | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, Nodulation fucosyltransferase NodZ, ... | | Authors: | Brzezinski, K, Dauter, Z, Jaskolski, M. | | Deposit date: | 2011-06-20 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of NodZ alpha-1,6-fucosyltransferase in complex with GDP and GDP-fucose
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3SIW
 
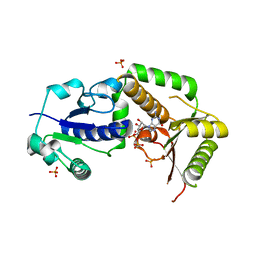 | | Crystal structure of NodZ alpha-1,6-fucosyltransferase co-crystallized with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Nodulation fucosyltransferase NodZ, PHOSPHATE ION | | Authors: | Brzezinski, K, Dauter, Z, Jaskolski, M. | | Deposit date: | 2011-06-20 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structures of NodZ alpha-1,6-fucosyltransferase in complex with GDP and GDP-fucose
Acta Crystallogr.,Sect.D, 68, 2012
|
|
